"steroid-5-alpha-reductase, alpha polypeptide 2a"
ZFIN 
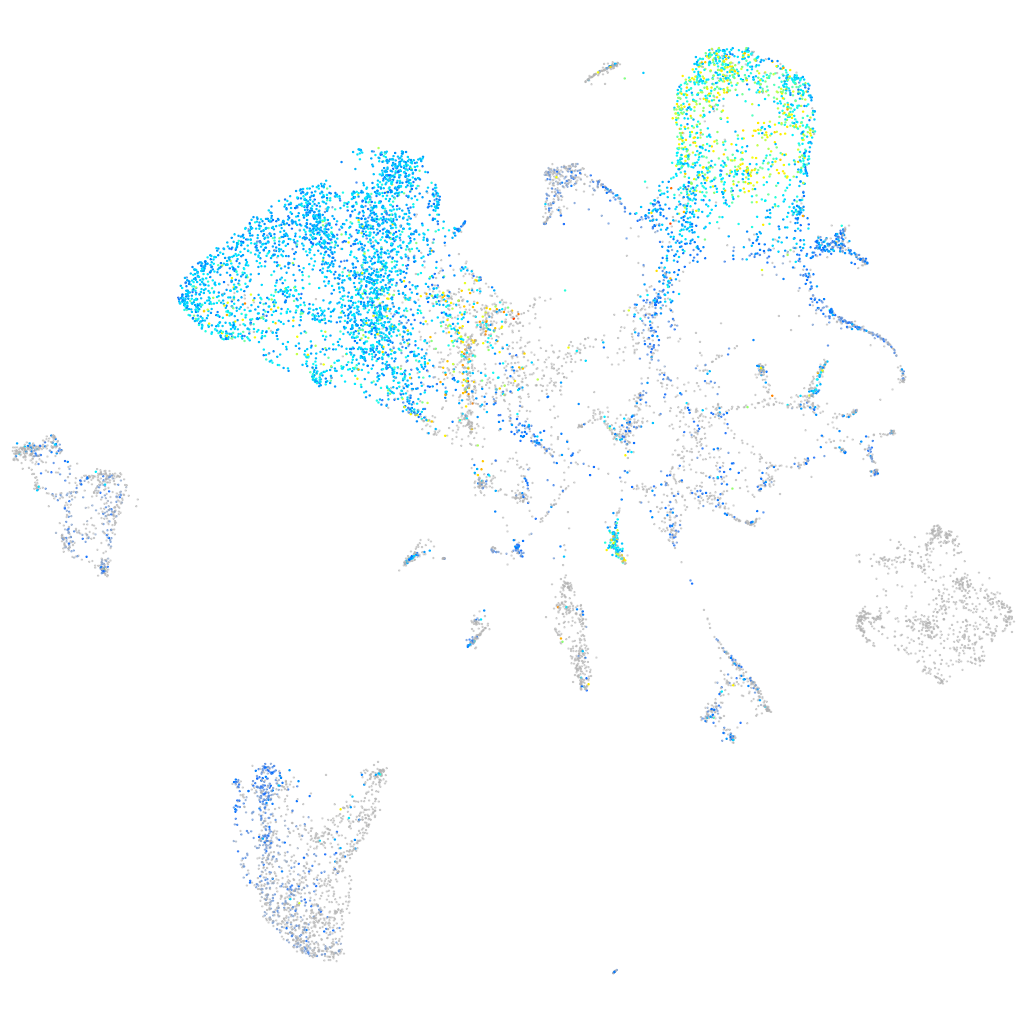
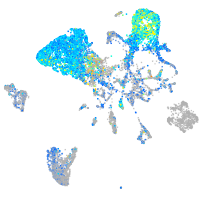
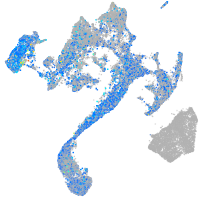
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scp2a | 0.702 | slc38a5b | -0.366 |
| apoa4b.1 | 0.698 | hmgb3a | -0.344 |
| rdh1 | 0.696 | rtn1a | -0.340 |
| dhrs9 | 0.692 | marcksl1b | -0.337 |
| glud1b | 0.690 | epcam | -0.335 |
| sult2st2 | 0.682 | hmgn6 | -0.330 |
| gcshb | 0.670 | glulb | -0.326 |
| apoc2 | 0.670 | marcksb | -0.326 |
| mdh1aa | 0.669 | si:ch211-222l21.1 | -0.318 |
| gpx4a | 0.668 | hmgb1b | -0.316 |
| suclg1 | 0.666 | si:ch211-195b11.3 | -0.309 |
| cyp8b1 | 0.666 | cx43.4 | -0.301 |
| sod1 | 0.661 | h3f3d | -0.300 |
| ugt1a7 | 0.659 | ywhaz | -0.299 |
| sdr16c5b | 0.655 | cldnb | -0.297 |
| eno3 | 0.654 | jpt1b | -0.297 |
| prdx2 | 0.650 | si:ch73-1a9.3 | -0.297 |
| gapdh | 0.645 | hspb1 | -0.290 |
| gstt1a | 0.644 | ctrl | -0.289 |
| gstr | 0.634 | cldn7b | -0.287 |
| slc37a4a | 0.627 | syncrip | -0.287 |
| fbp1b | 0.622 | eef1a1l1 | -0.286 |
| pgam1a | 0.621 | id1 | -0.286 |
| ugt5b4 | 0.621 | akap12b | -0.282 |
| tpi1b | 0.620 | nucks1a | -0.280 |
| fdx1 | 0.619 | bcam | -0.278 |
| mttp | 0.616 | hmgb2a | -0.277 |
| atp5mc1 | 0.611 | hmgb1a | -0.275 |
| slco1d1 | 0.610 | hnrnpa1a | -0.275 |
| mgst1.2 | 0.604 | acin1a | -0.274 |
| pgk1 | 0.600 | qkia | -0.274 |
| atp5f1b | 0.596 | hnrnpa0a | -0.274 |
| apobb.1 | 0.593 | tuba8l4 | -0.273 |
| afp4 | 0.593 | zgc:112160 | -0.273 |
| haao | 0.589 | ela3l | -0.273 |