spindlin a
ZFIN 
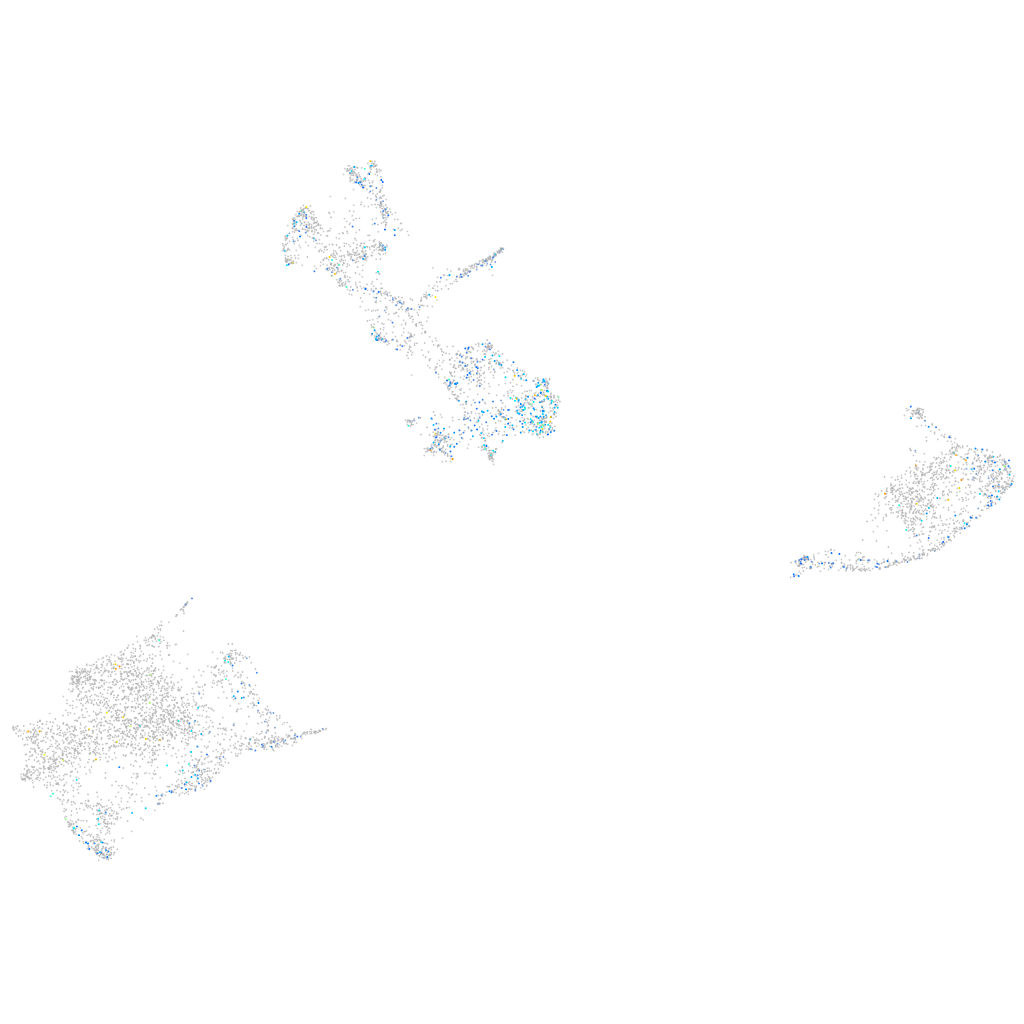
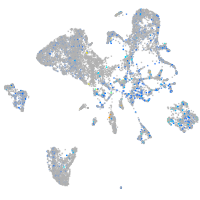
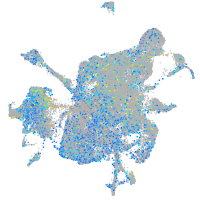
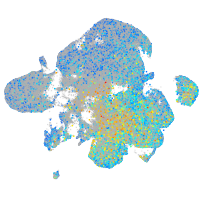
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gpm6aa | 0.207 | gch2 | -0.105 |
| zc4h2 | 0.187 | pts | -0.097 |
| nova2 | 0.187 | akr1b1 | -0.096 |
| stmn1b | 0.186 | si:dkey-151g10.6 | -0.090 |
| gng3 | 0.186 | gstp1 | -0.088 |
| elavl3 | 0.185 | paics | -0.086 |
| pbx1a | 0.183 | si:dkey-251i10.2 | -0.085 |
| rtn1a | 0.182 | CABZ01021592.1 | -0.085 |
| rnasekb | 0.181 | slc22a7a | -0.083 |
| elavl4 | 0.178 | actb2 | -0.079 |
| fez1 | 0.176 | zgc:110339 | -0.079 |
| tmeff1b | 0.174 | pfn1 | -0.076 |
| atp6v0cb | 0.172 | tmem130 | -0.075 |
| gnao1a | 0.170 | uraha | -0.072 |
| hmgb1b | 0.167 | gpd1b | -0.070 |
| stx1b | 0.166 | pcbd1 | -0.069 |
| tubb5 | 0.166 | rps2 | -0.064 |
| sncb | 0.166 | cyb5a | -0.064 |
| celf3a | 0.163 | sprb | -0.061 |
| cadm3 | 0.162 | prdx1 | -0.059 |
| si:dkey-276j7.1 | 0.161 | prdx5 | -0.055 |
| gng2 | 0.161 | bco1 | -0.054 |
| nova1 | 0.159 | krt18b | -0.054 |
| gpr85 | 0.158 | cmtm3 | -0.053 |
| pbx3b | 0.158 | impdh1b | -0.052 |
| fam168a | 0.156 | atic | -0.052 |
| ankrd12 | 0.155 | rbp4l | -0.051 |
| zfhx3 | 0.154 | aqp3a | -0.050 |
| hmgb3a | 0.152 | rplp0 | -0.049 |
| tmpob | 0.152 | cx30.3 | -0.048 |
| celf4 | 0.152 | rpl12 | -0.048 |
| id4 | 0.150 | CABZ01032488.1 | -0.048 |
| tuba1c | 0.150 | aldob | -0.047 |
| cnrip1a | 0.149 | sult1st1 | -0.047 |
| si:dkey-56f14.7 | 0.148 | TMEM19 | -0.046 |