"SPG21 abhydrolase domain containing, maspardin"
ZFIN 
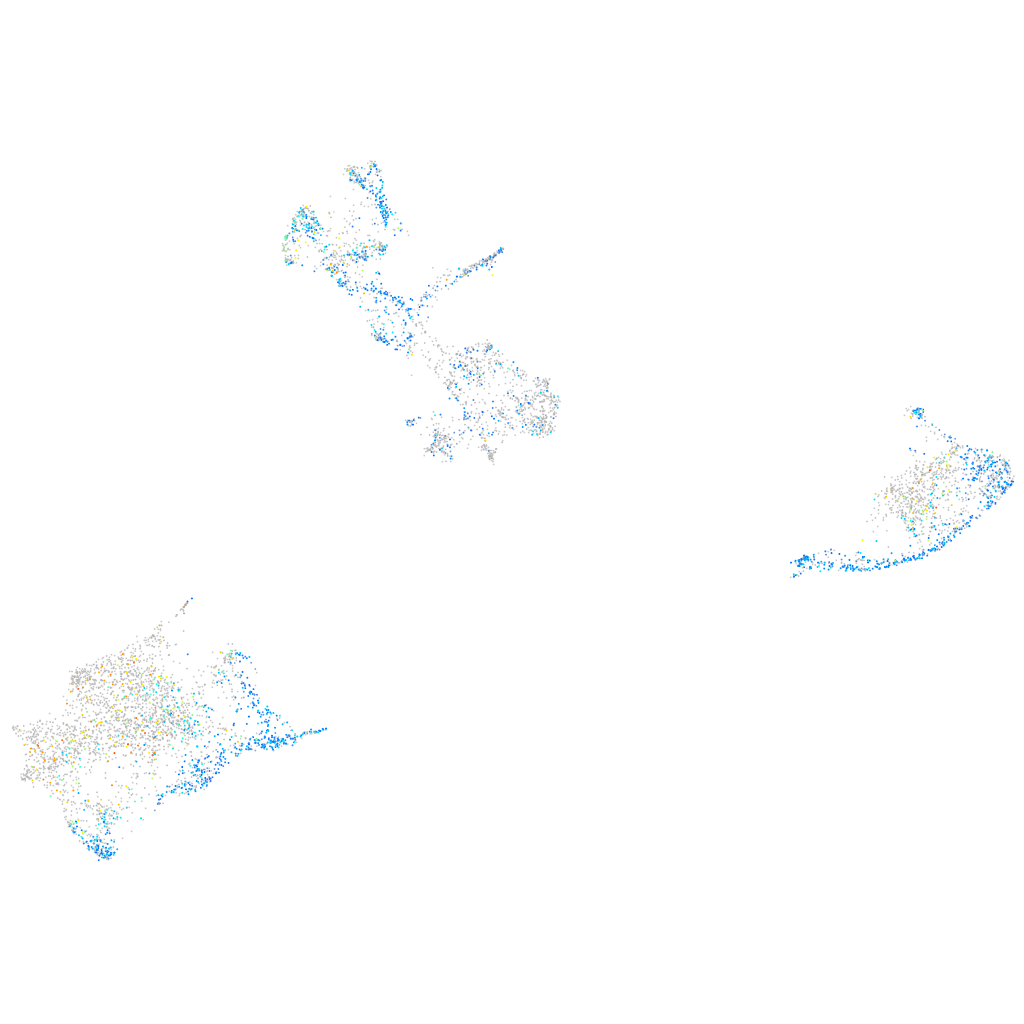
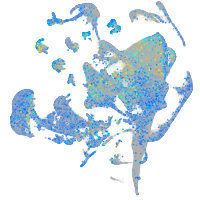
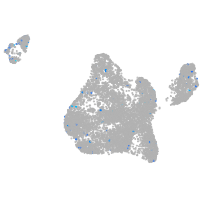
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| zgc:110239 | 0.183 | hbbe1.3 | -0.112 |
| rnaseka | 0.182 | tuba1c | -0.112 |
| ctsba | 0.182 | pvalb1 | -0.110 |
| trpm1b | 0.180 | pvalb2 | -0.108 |
| inka1a | 0.174 | ptmaa | -0.106 |
| smim29 | 0.174 | gpm6aa | -0.104 |
| rab32a | 0.173 | myhz1.1 | -0.101 |
| gpr143 | 0.173 | hbae3 | -0.098 |
| syngr1a | 0.172 | nova2 | -0.096 |
| si:zfos-943e10.1 | 0.168 | stmn1b | -0.096 |
| atox1 | 0.166 | tmsb | -0.095 |
| sdc4 | 0.163 | elavl3 | -0.094 |
| npm1a | 0.161 | mylpfa | -0.093 |
| sypl2b | 0.160 | actc1b | -0.091 |
| rgs3a | 0.160 | sncb | -0.086 |
| agtrap | 0.160 | hbae1.1 | -0.086 |
| BX571715.1 | 0.159 | si:ch73-1a9.3 | -0.086 |
| lamtor5 | 0.159 | CU467822.1 | -0.086 |
| hspe1 | 0.159 | sox11b | -0.083 |
| pnp4a | 0.157 | gng3 | -0.083 |
| pttg1ipb | 0.157 | si:ch211-222l21.1 | -0.083 |
| sptlc2a | 0.157 | krt4 | -0.083 |
| tfec | 0.156 | fabp7a | -0.082 |
| cst14a.2 | 0.155 | epb41a | -0.080 |
| adi1 | 0.153 | hbbe1.1 | -0.080 |
| zgc:165573 | 0.153 | tox | -0.079 |
| bloc1s4 | 0.152 | cdh2 | -0.078 |
| dtnbp1a | 0.152 | zc4h2 | -0.077 |
| gadd45bb | 0.152 | tnnt3b | -0.075 |
| dera | 0.150 | gng2 | -0.074 |
| rnaset2 | 0.149 | cyt1 | -0.074 |
| hexb | 0.149 | vamp2 | -0.073 |
| lamtor2 | 0.149 | mylz3 | -0.073 |
| lman2 | 0.146 | cadm3 | -0.072 |
| adsl | 0.146 | tuba1a | -0.072 |