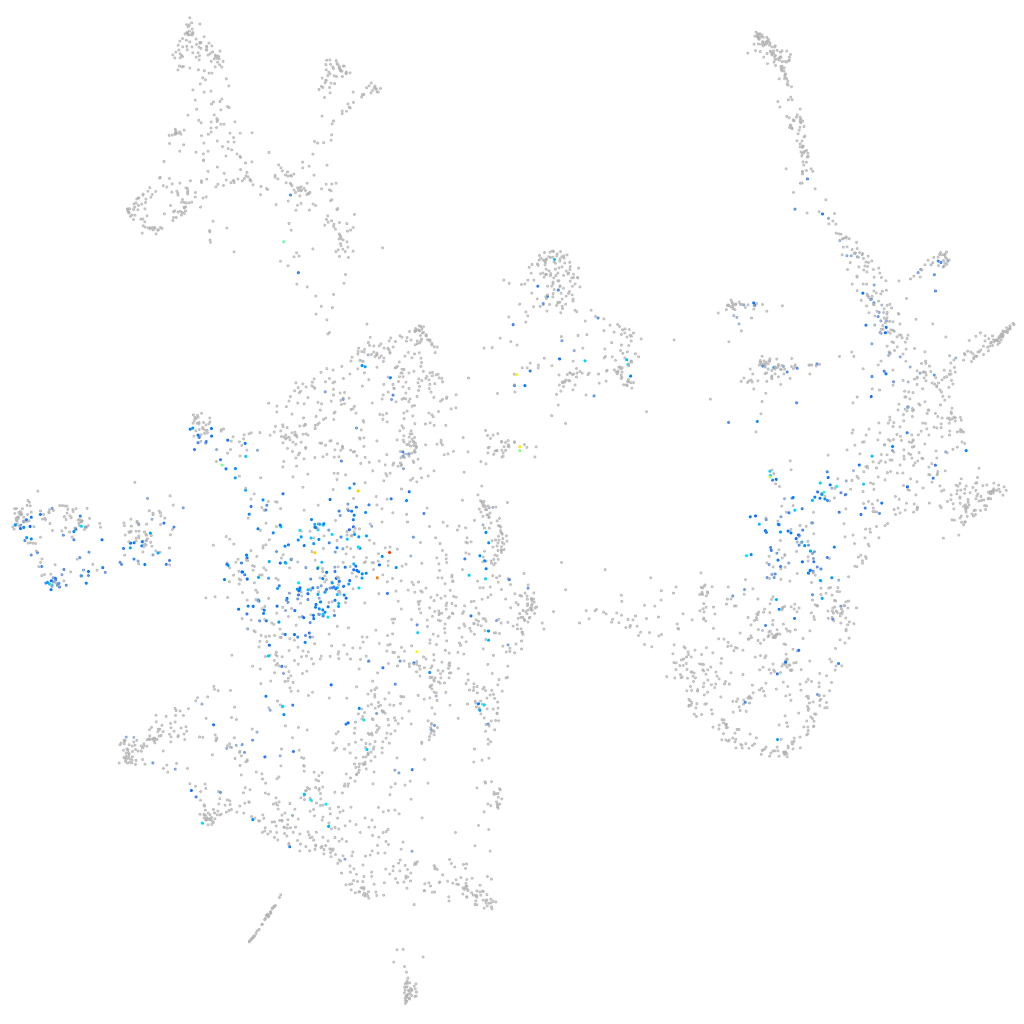
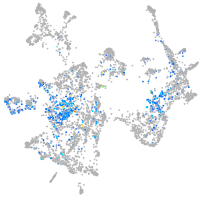
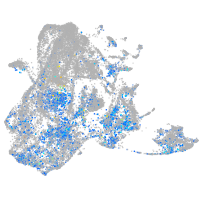
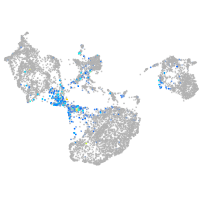
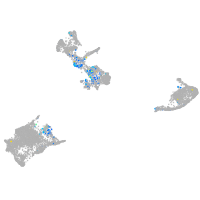
SPC24 component of NDC80 kinetochore complex
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cks1b | 0.516 | tob1b | -0.158 |
| mad2l1 | 0.513 | COX3 | -0.139 |
| mki67 | 0.500 | atp6v1e1b | -0.128 |
| ccna2 | 0.497 | mt-co2 | -0.126 |
| cdk1 | 0.495 | atp2b1a | -0.125 |
| top2a | 0.468 | b2ml | -0.124 |
| aurkb | 0.458 | calml4a | -0.123 |
| plk1 | 0.453 | otofb | -0.122 |
| ube2c | 0.453 | nptna | -0.121 |
| tpx2 | 0.448 | tmem59 | -0.121 |
| kif23 | 0.443 | ccni | -0.120 |
| cdc20 | 0.437 | gpx2 | -0.119 |
| ttk | 0.425 | atp1a3b | -0.119 |
| cdca5 | 0.421 | gabarapl2 | -0.118 |
| ncapg | 0.418 | cd164l2 | -0.118 |
| cenpf | 0.418 | eno1a | -0.118 |
| zgc:165555.3 | 0.418 | calm1a | -0.117 |
| kifc1 | 0.417 | calm1b | -0.117 |
| nusap1 | 0.417 | osbpl1a | -0.116 |
| dlgap5 | 0.415 | si:ch73-15n15.3 | -0.114 |
| spdl1 | 0.414 | CR925719.1 | -0.114 |
| hmgb2a | 0.410 | si:dkey-16p21.8 | -0.113 |
| ccnb1 | 0.409 | gabarapb | -0.113 |
| cdca8 | 0.407 | gabarapa | -0.113 |
| aspm | 0.406 | tspan13b | -0.113 |
| aurka | 0.406 | pvalb8 | -0.113 |
| armc1l | 0.403 | ap3s1.1 | -0.113 |
| kif11 | 0.403 | pgk1 | -0.112 |
| spc25 | 0.400 | gapdhs | -0.112 |
| birc5a | 0.400 | kif1aa | -0.112 |
| cenpe | 0.399 | pkig | -0.111 |
| stmn1a | 0.397 | nupr1a | -0.110 |
| prc1a | 0.396 | dnajc5b | -0.110 |
| kpna2 | 0.395 | lrrc73 | -0.110 |
| mibp | 0.389 | ckbb | -0.109 |