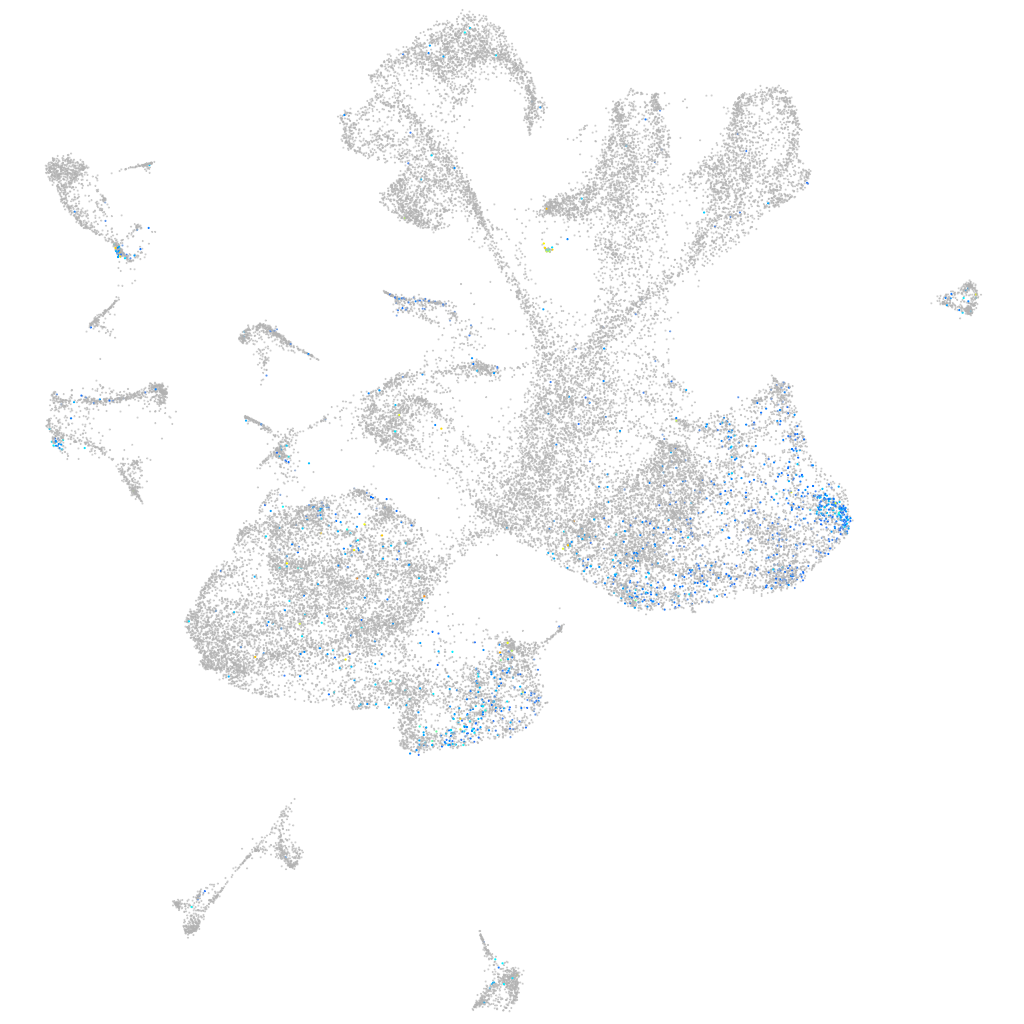
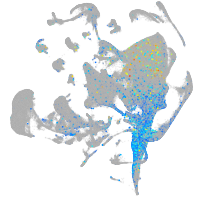
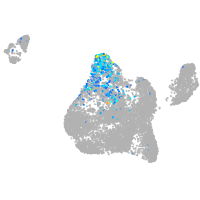
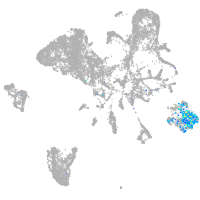
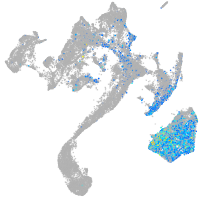
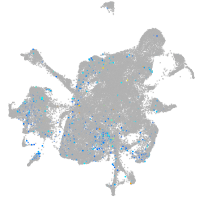
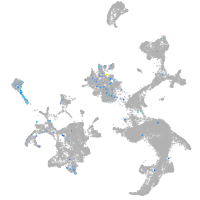
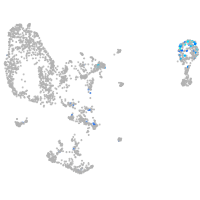
Sp5 transcription factor a
ZFIN 
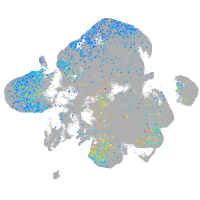
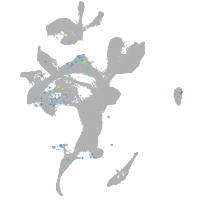
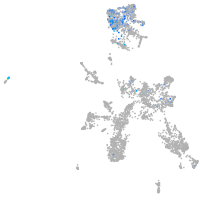
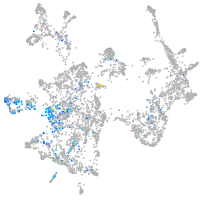
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| noto | 0.224 | ckbb | -0.086 |
| sp5l | 0.180 | elavl3 | -0.084 |
| fgf8a | 0.161 | gpm6aa | -0.083 |
| cdx1a | 0.159 | myt1b | -0.078 |
| nradd | 0.159 | rtn1a | -0.078 |
| znfl2a | 0.158 | tmsb | -0.076 |
| chrd | 0.150 | vim | -0.072 |
| cdx4 | 0.141 | pvalb1 | -0.072 |
| apela | 0.137 | tmsb4x | -0.071 |
| pou5f3 | 0.136 | stmn1b | -0.071 |
| nkx1.2la | 0.136 | pvalb2 | -0.069 |
| pcna | 0.134 | rnasekb | -0.068 |
| hspb1 | 0.134 | marcksl1b | -0.068 |
| foxd5 | 0.129 | si:dkeyp-75h12.5 | -0.066 |
| cxcr4a | 0.129 | elavl4 | -0.064 |
| chaf1a | 0.124 | hbbe1.3 | -0.063 |
| nasp | 0.124 | fabp3 | -0.061 |
| rpa2 | 0.123 | actc1b | -0.061 |
| stmn1a | 0.121 | CR383676.1 | -0.061 |
| npm1a | 0.120 | gapdhs | -0.059 |
| mcm7 | 0.119 | gpm6ab | -0.059 |
| dek | 0.119 | hbae3 | -0.059 |
| dut | 0.118 | stxbp1a | -0.059 |
| ccnd1 | 0.117 | scrt2 | -0.059 |
| slbp | 0.116 | gnao1a | -0.058 |
| fen1 | 0.116 | sncb | -0.056 |
| lrrc17 | 0.114 | COX3 | -0.056 |
| mcm6 | 0.113 | calm1a | -0.056 |
| rrm2 | 0.113 | cplx2 | -0.056 |
| anp32b | 0.113 | cadm3 | -0.056 |
| ranbp1 | 0.112 | syt2a | -0.056 |
| tbxta | 0.112 | slc1a2b | -0.055 |
| nutf2l | 0.112 | nova2 | -0.055 |
| lig1 | 0.111 | onecut1 | -0.055 |
| selenoh | 0.110 | gng3 | -0.055 |