SRY-box transcription factor 7
ZFIN 
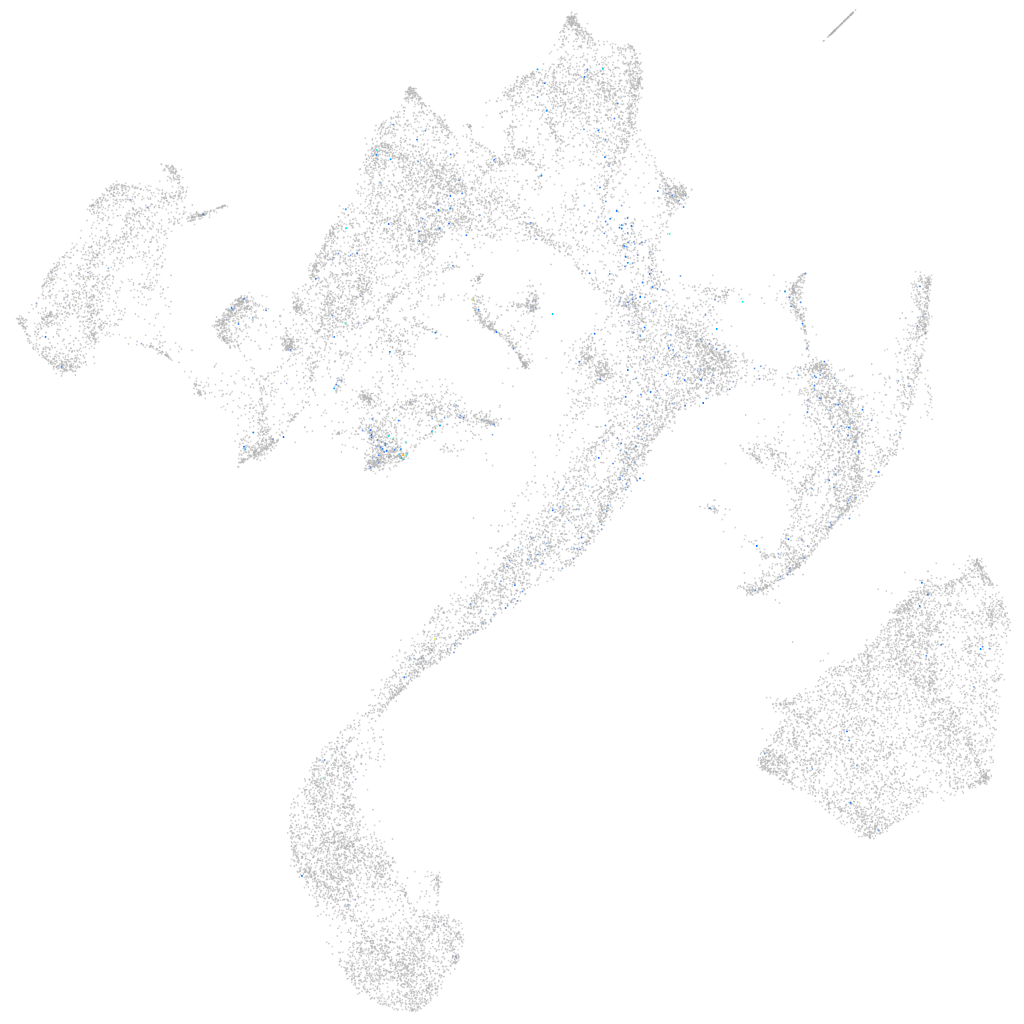
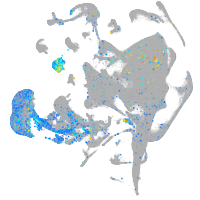
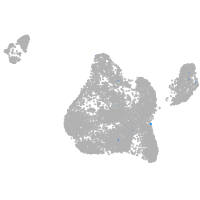
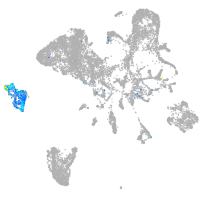
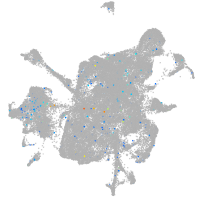
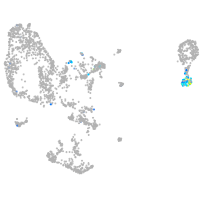
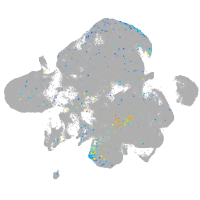
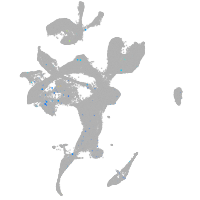
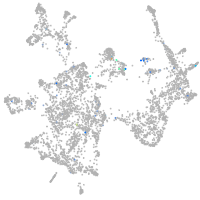
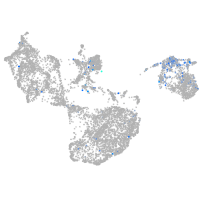
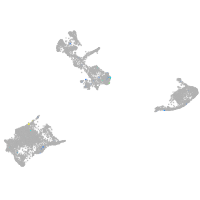
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pecam1 | 0.199 | actn3a | -0.038 |
| rasip1 | 0.182 | XLOC-025819 | -0.036 |
| clec14a | 0.179 | casq1b | -0.036 |
| abcb4 | 0.155 | tmod4 | -0.035 |
| fli1b | 0.155 | ldb3b | -0.035 |
| cdh5 | 0.154 | si:ch211-255p10.3 | -0.035 |
| LOC103910788 | 0.136 | smyd1a | -0.034 |
| she | 0.129 | ckmb | -0.034 |
| kdr | 0.125 | si:ch211-266g18.10 | -0.034 |
| sult2st3 | 0.115 | prx | -0.034 |
| tie1 | 0.115 | si:ch211-152c2.3 | -0.034 |
| shank3b | 0.114 | pgam2 | -0.034 |
| plvapb | 0.109 | actn3b | -0.034 |
| XLOC-014079 | 0.106 | XLOC-005350 | -0.033 |
| znf997 | 0.104 | neb | -0.033 |
| scrt1a | 0.102 | hhatla | -0.033 |
| bcl6b | 0.101 | XLOC-006515 | -0.033 |
| hhex | 0.099 | pkmb | -0.032 |
| XLOC-043043 | 0.097 | myom1a | -0.032 |
| tnfaip2b | 0.095 | myom1b | -0.032 |
| tal1 | 0.094 | si:ch73-367p23.2 | -0.032 |
| etv2 | 0.094 | CR855257.1 | -0.031 |
| CR388042.2 | 0.093 | gapdh | -0.031 |
| arhgef9b | 0.087 | trdn | -0.031 |
| gpr182 | 0.084 | glud1b | -0.031 |
| lmo2 | 0.083 | si:dkey-211f22.5 | -0.031 |
| si:dkey-47k16.4 | 0.083 | unm-hu7910 | -0.031 |
| BX323067.1 | 0.080 | dhrs7cb | -0.031 |
| fgd5a | 0.076 | jph1b | -0.031 |
| pcdh12 | 0.076 | casq2 | -0.031 |
| bdkrb1 | 0.075 | cox7c | -0.030 |
| onecut1 | 0.075 | mylpfb | -0.030 |
| elavl3 | 0.075 | BX927327.1 | -0.030 |
| lhx9 | 0.074 | cdx1a | -0.030 |
| CU041345.1 | 0.074 | cdx4 | -0.030 |