SRY-box transcription factor 3
ZFIN 
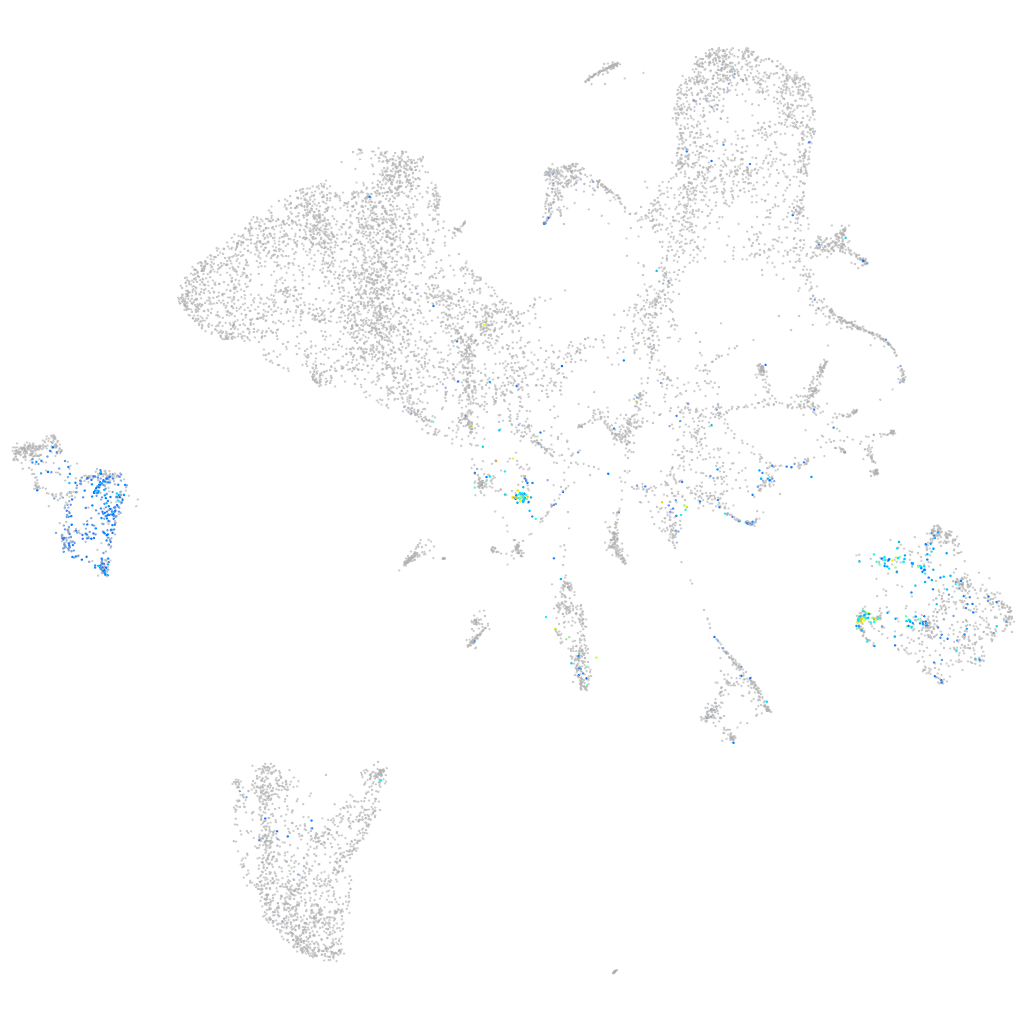
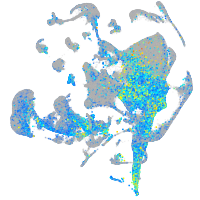
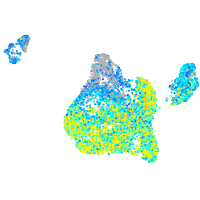
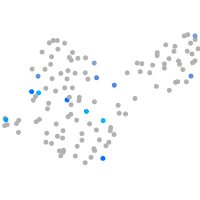
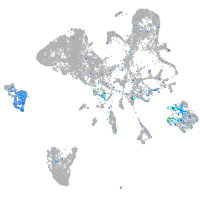
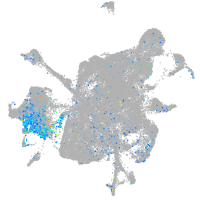
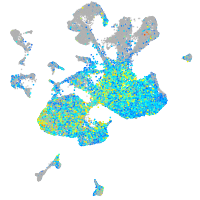
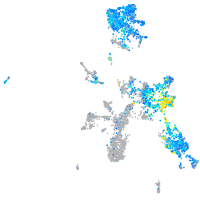
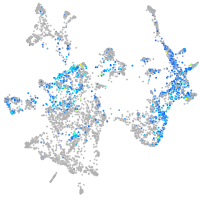
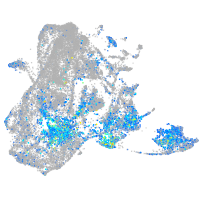
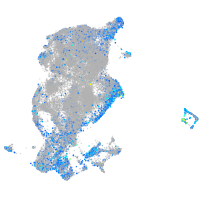
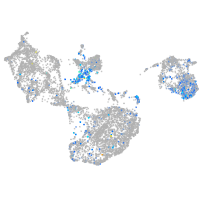
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| sox19a | 0.325 | gapdh | -0.245 |
| XLOC-003690 | 0.318 | eno3 | -0.217 |
| rarga | 0.315 | pklr | -0.210 |
| XLOC-003689 | 0.291 | gamt | -0.200 |
| sp8b | 0.278 | suclg1 | -0.196 |
| cldn5a | 0.278 | glud1b | -0.191 |
| mdka | 0.273 | ahcy | -0.189 |
| gbx2 | 0.261 | lgals2b | -0.188 |
| apela | 0.256 | tpi1b | -0.183 |
| apoeb | 0.256 | rpl37 | -0.183 |
| hmgb1b | 0.255 | gstt1a | -0.181 |
| sox2 | 0.255 | fbp1b | -0.181 |
| mir219-3 | 0.252 | scp2a | -0.180 |
| hmgn6 | 0.252 | gpx4a | -0.179 |
| id1 | 0.249 | cx32.3 | -0.179 |
| hmgb2a | 0.248 | dap | -0.178 |
| nkx2.7 | 0.244 | mat1a | -0.177 |
| ptmab | 0.242 | rps10 | -0.176 |
| si:ch211-137a8.4 | 0.241 | sod1 | -0.175 |
| hspb1 | 0.239 | cox6b1 | -0.175 |
| si:ch73-281n10.2 | 0.238 | nupr1b | -0.175 |
| si:ch211-152c2.3 | 0.237 | BX908782.3 | -0.174 |
| msi1 | 0.236 | atp5mc1 | -0.174 |
| syncrip | 0.235 | gatm | -0.174 |
| cx43.4 | 0.234 | prdx2 | -0.171 |
| si:ch73-1a9.3 | 0.234 | aldob | -0.170 |
| seta | 0.233 | zgc:92744 | -0.169 |
| marcksb | 0.233 | apoa4b.1 | -0.168 |
| cxcl12b | 0.233 | gcshb | -0.165 |
| cdkn1ca | 0.233 | aldh7a1 | -0.164 |
| serpinh1b | 0.232 | mdh1aa | -0.164 |
| pltp | 0.231 | zgc:114188 | -0.164 |
| h3f3d | 0.231 | abat | -0.164 |
| si:ch211-222l21.1 | 0.230 | apoc2 | -0.164 |
| XLOC-003692 | 0.228 | pgk1 | -0.163 |