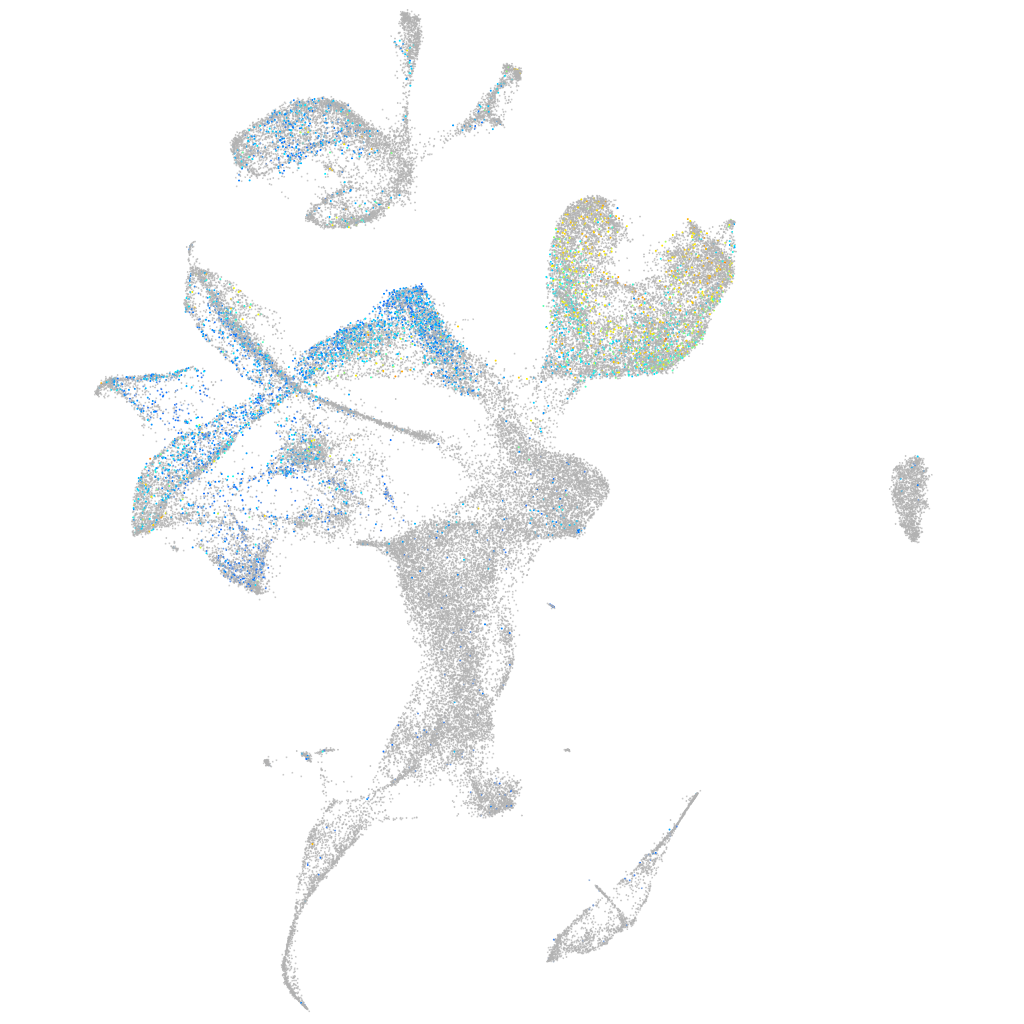sosondowah ankyrin repeat domain family Cb
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| crx | 0.202 | zgc:56493 | -0.126 |
| syt5b | 0.177 | her15.1 | -0.126 |
| otx5 | 0.177 | pcna | -0.123 |
| ckbb | 0.174 | mdka | -0.121 |
| sypb | 0.165 | ranbp1 | -0.121 |
| rs1a | 0.155 | tspan7 | -0.120 |
| atp6v0cb | 0.146 | id1 | -0.118 |
| anp32e | 0.142 | rrm1 | -0.117 |
| snap25b | 0.141 | cldn5a | -0.116 |
| zgc:109965 | 0.139 | mcm7 | -0.116 |
| mpp6b | 0.139 | rpl9 | -0.116 |
| gngt2a | 0.138 | si:ch211-222l21.1 | -0.115 |
| neurod1 | 0.138 | tuba8l | -0.110 |
| atp2b1b | 0.135 | banf1 | -0.109 |
| rnasekb | 0.135 | msna | -0.109 |
| atpv0e2 | 0.133 | XLOC-003690 | -0.107 |
| si:ch73-28h20.1 | 0.132 | ccnd1 | -0.107 |
| atp1a3a | 0.132 | COX7A2 (1 of many) | -0.107 |
| pdcl | 0.131 | mcm6 | -0.107 |
| elovl4b | 0.130 | selenoh | -0.106 |
| gapdhs | 0.130 | nasp | -0.104 |
| ddah2 | 0.130 | lig1 | -0.104 |
| epb41a | 0.130 | cdca7b | -0.103 |
| tulp1a | 0.129 | eif5a2 | -0.103 |
| gpm6ab | 0.127 | anp32b | -0.102 |
| opn6a | 0.126 | nutf2l | -0.102 |
| tmx3a | 0.125 | hes6 | -0.102 |
| cnrip1b | 0.124 | mcm5 | -0.102 |
| arl3l1 | 0.123 | mcm2 | -0.101 |
| arl13a | 0.122 | sox2 | -0.101 |
| scg3 | 0.122 | tuba8l4 | -0.100 |
| xbp1 | 0.122 | ahcy | -0.100 |
| zgc:103625 | 0.122 | phgdh | -0.100 |
| stxbp1b | 0.121 | rrs1 | -0.099 |
| gnb3a | 0.121 | cdca7a | -0.098 |