SET and MYND domain containing 2a
ZFIN 
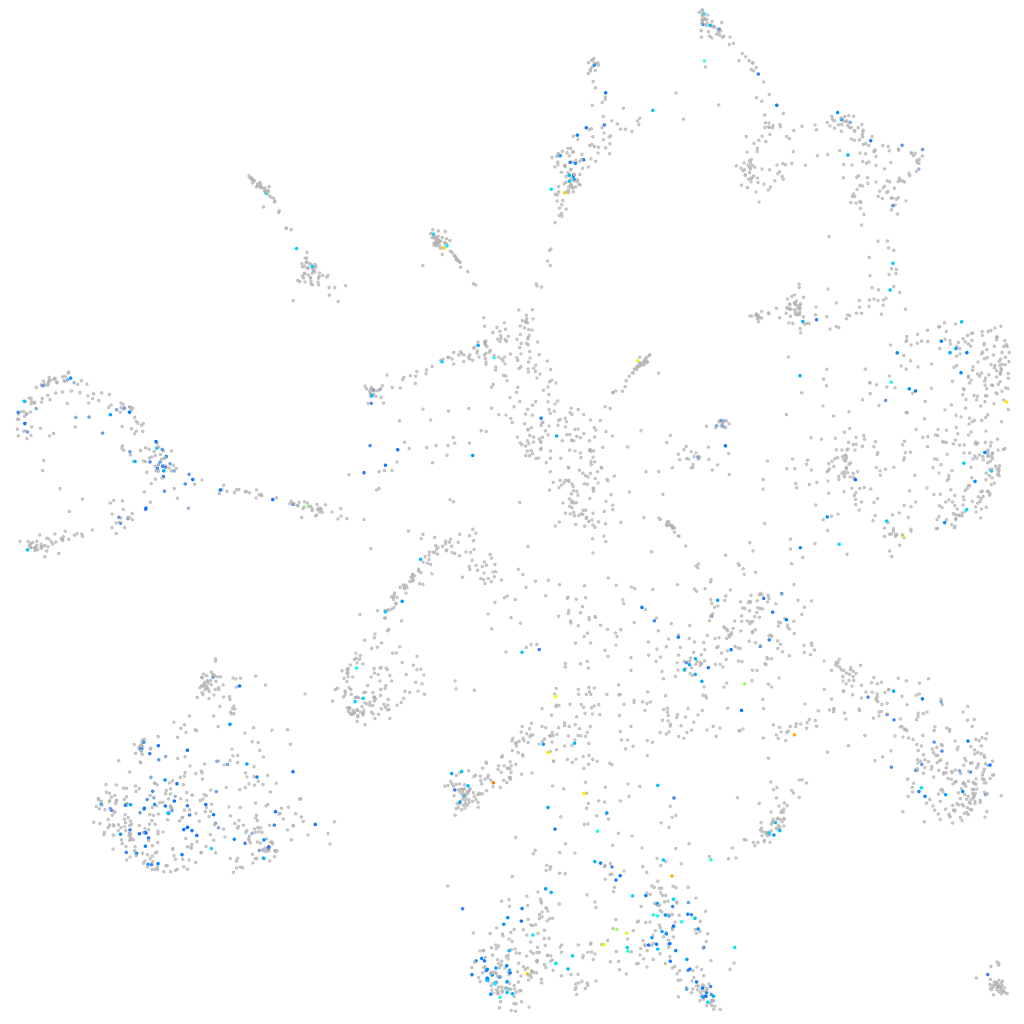
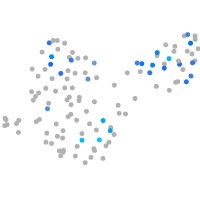
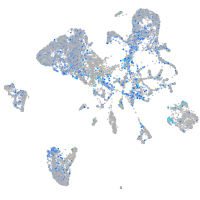
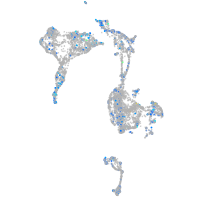
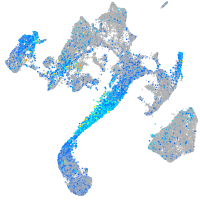
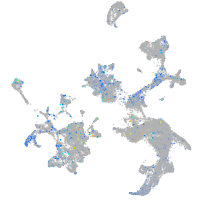
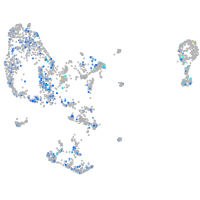
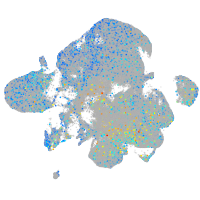
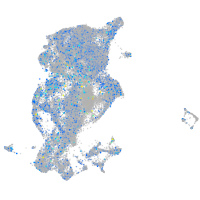
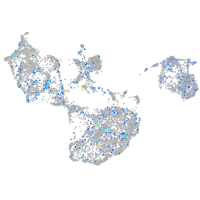
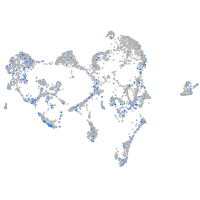
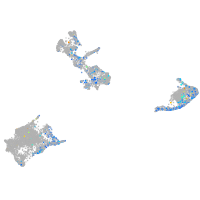
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cald1b | 0.195 | si:ch1073-291c23.2 | -0.078 |
| tpm2 | 0.175 | igf2b | -0.073 |
| fbxl22 | 0.174 | rbp4 | -0.072 |
| csrp1b | 0.171 | ednraa | -0.071 |
| desmb | 0.161 | cxcl12a | -0.070 |
| cnn1b | 0.161 | cd248a | -0.070 |
| fhl1a | 0.160 | histh1l | -0.065 |
| myh11a | 0.159 | id1 | -0.064 |
| mylkb | 0.155 | CR383676.1 | -0.062 |
| acta1b | 0.151 | cox4i2 | -0.061 |
| pdlim3a | 0.151 | lsp1 | -0.059 |
| myl6 | 0.150 | si:dkey-261h17.1 | -0.059 |
| trim35-9 | 0.150 | cebpa | -0.057 |
| acta2 | 0.148 | si:dkey-238c7.16 | -0.057 |
| myl9a | 0.146 | si:dkey-9i23.16 | -0.056 |
| smtna | 0.145 | notch3 | -0.055 |
| tuba8l2 | 0.142 | trim69 | -0.054 |
| si:ch211-137i24.10 | 0.142 | c1qtnf6a | -0.054 |
| tagln | 0.141 | tmem88b | -0.054 |
| lmod1b | 0.133 | nrp1a | -0.053 |
| tpm1 | 0.130 | rbpms2b | -0.053 |
| myocd | 0.130 | tp53inp1 | -0.053 |
| acta1a | 0.129 | mknk2b | -0.052 |
| pnp4a | 0.128 | ecm1b | -0.052 |
| ckba | 0.127 | podxl | -0.052 |
| ckbb | 0.126 | pvalb1 | -0.050 |
| npb | 0.125 | zeb2a | -0.050 |
| CABZ01056629.1 | 0.123 | cebpb | -0.050 |
| vcla | 0.123 | bambia | -0.050 |
| cox6b1 | 0.122 | si:ch211-160j14.3 | -0.049 |
| kcnk18 | 0.120 | tbx2b | -0.049 |
| pdlim3b | 0.120 | eva1bb | -0.049 |
| gucy1a1 | 0.119 | cxcl12b | -0.048 |
| RF00263 | 0.119 | sparc | -0.048 |
| itpka | 0.117 | fabp11a | -0.048 |