small integral membrane protein 20
ZFIN 
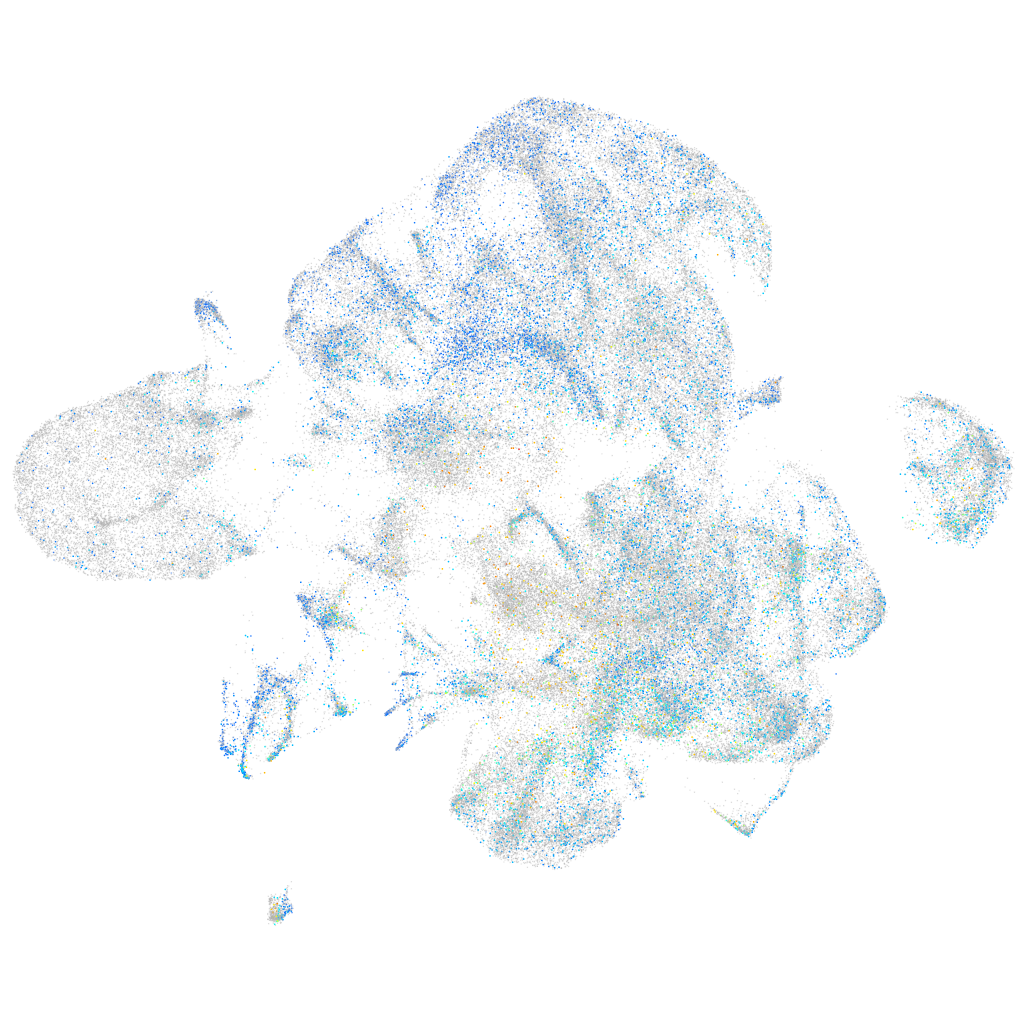
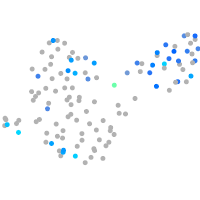
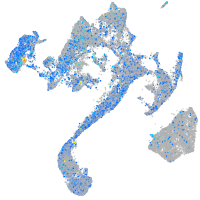
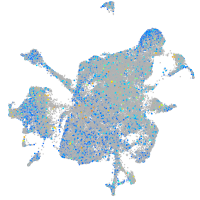
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gapdhs | 0.101 | NC-002333.4 | -0.065 |
| atp5mc1 | 0.095 | si:ch211-152c2.3 | -0.061 |
| cox7a2a | 0.089 | stm | -0.059 |
| tpi1b | 0.089 | hspb1 | -0.058 |
| calm1b | 0.087 | hmga1a | -0.054 |
| atp6v1e1b | 0.084 | si:ch211-222l21.1 | -0.052 |
| atp5f1b | 0.082 | pou5f3 | -0.051 |
| calm3a | 0.081 | crabp2b | -0.050 |
| atp6v1g1 | 0.081 | apoeb | -0.050 |
| ppiab | 0.080 | polr3gla | -0.049 |
| rnasekb | 0.079 | apela | -0.048 |
| aldocb | 0.078 | akap12b | -0.047 |
| atp5l | 0.078 | zmp:0000000624 | -0.046 |
| ndufa4 | 0.077 | s100a1 | -0.046 |
| si:ch73-119p20.1 | 0.077 | apoc1 | -0.045 |
| atp6v0cb | 0.077 | si:dkey-66i24.9 | -0.045 |
| eno1a | 0.076 | cx43.4 | -0.045 |
| ndrg3a | 0.076 | shisa2a | -0.045 |
| calm3b | 0.076 | lig1 | -0.044 |
| calm1a | 0.076 | dnmt3bb.2 | -0.043 |
| ldhba | 0.076 | sfrp1a | -0.043 |
| cox8a | 0.076 | hnrnpa1b | -0.042 |
| atp5meb | 0.075 | hmgb2a | -0.042 |
| zgc:65894 | 0.075 | COX7A2 | -0.042 |
| cox6a1 | 0.074 | dkc1 | -0.042 |
| pkma | 0.074 | bms1 | -0.042 |
| atp6ap2 | 0.074 | nr6a1a | -0.042 |
| cox4i1 | 0.074 | rsl1d1 | -0.042 |
| rab6bb | 0.074 | pkdccb | -0.042 |
| cdc42 | 0.074 | fbl | -0.042 |
| atp5if1b | 0.074 | nop58 | -0.042 |
| nptna | 0.073 | nop56 | -0.041 |
| sh3gl2a | 0.073 | zgc:110425 | -0.041 |
| stmn2a | 0.073 | asb11 | -0.041 |
| sncgb | 0.073 | rps18 | -0.041 |