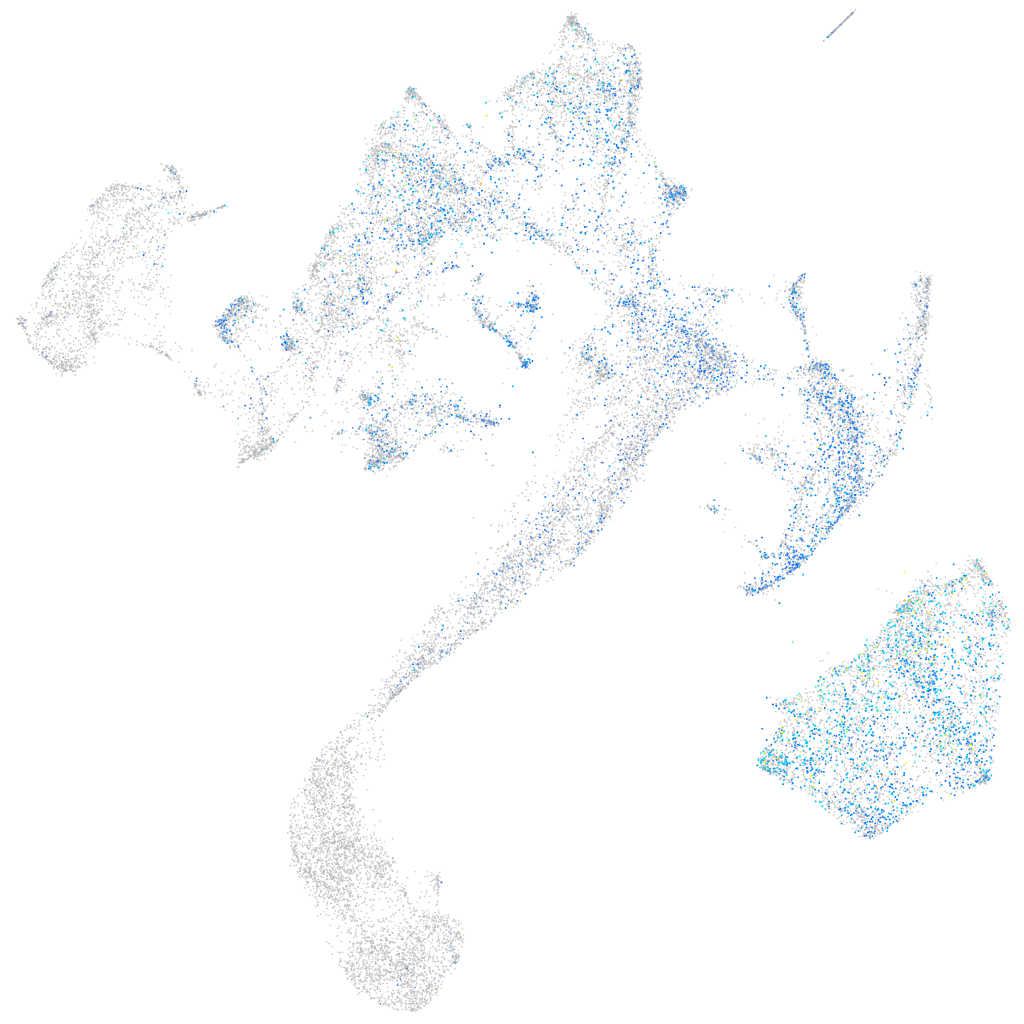
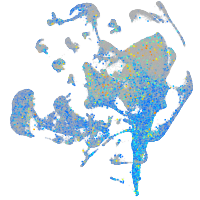
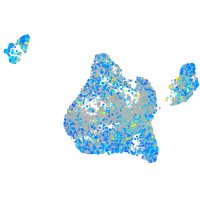
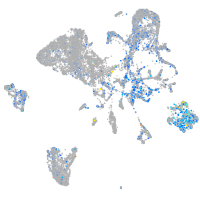
structural maintenance of chromosomes 5
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| chaf1a | 0.283 | actc1b | -0.249 |
| lig1 | 0.281 | rpl37 | -0.228 |
| seta | 0.275 | ttn.2 | -0.228 |
| cx43.4 | 0.272 | ttn.1 | -0.222 |
| si:ch211-222l21.1 | 0.272 | ak1 | -0.220 |
| ptmab | 0.271 | ckmb | -0.219 |
| hnrnpa1b | 0.268 | atp2a1 | -0.219 |
| nucks1a | 0.266 | ckma | -0.218 |
| acin1a | 0.265 | aldoab | -0.216 |
| anp32b | 0.263 | tmem38a | -0.211 |
| cdx4 | 0.263 | tnnc2 | -0.210 |
| hmgb2a | 0.262 | zgc:114188 | -0.208 |
| cirbpa | 0.261 | rps10 | -0.206 |
| si:ch73-1a9.3 | 0.261 | neb | -0.202 |
| hmga1a | 0.261 | acta1b | -0.202 |
| marcksb | 0.260 | mylpfa | -0.200 |
| nasp | 0.259 | pabpc4 | -0.200 |
| banf1 | 0.258 | atp5meb | -0.200 |
| anp32e | 0.258 | mybphb | -0.200 |
| anp32a | 0.257 | tpma | -0.198 |
| hnrnpub | 0.256 | ldb3a | -0.195 |
| dek | 0.254 | nme2b.2 | -0.195 |
| ppm1g | 0.254 | rps17 | -0.195 |
| hnrnpa1a | 0.252 | CABZ01078594.1 | -0.195 |
| pcna | 0.250 | srl | -0.194 |
| pabpc1a | 0.249 | desma | -0.194 |
| baz1b | 0.249 | mdh2 | -0.192 |
| si:ch73-281n10.2 | 0.248 | ldb3b | -0.192 |
| ppig | 0.247 | actn3b | -0.192 |
| hmgn7 | 0.247 | gapdh | -0.191 |
| smc1al | 0.246 | atp5if1b | -0.191 |
| hnrnpaba | 0.243 | cav3 | -0.190 |
| ncl | 0.243 | gamt | -0.190 |
| syncrip | 0.243 | eno3 | -0.190 |
| hirip3 | 0.242 | si:ch73-367p23.2 | -0.189 |