structural maintenance of chromosomes 3
ZFIN 
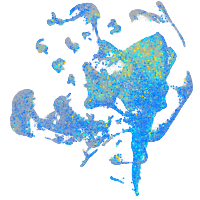
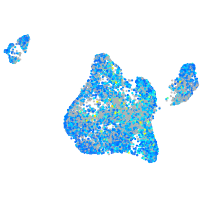
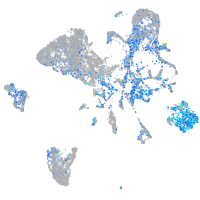
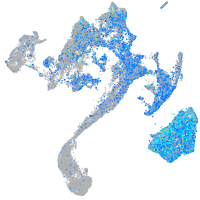
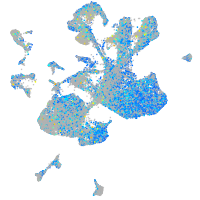
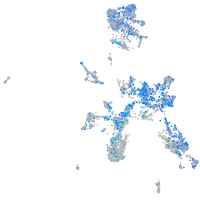
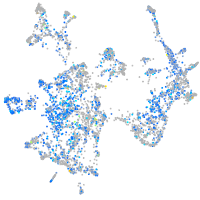
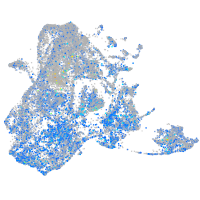
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| nucks1a | 0.444 | gamt | -0.317 |
| hnrnpa1b | 0.439 | rpl37 | -0.295 |
| hmga1a | 0.436 | rps10 | -0.287 |
| seta | 0.434 | gapdh | -0.286 |
| hnrnpub | 0.427 | gatm | -0.281 |
| hnrnpa1a | 0.426 | bhmt | -0.273 |
| hmgb2a | 0.422 | ahcy | -0.272 |
| syncrip | 0.418 | zgc:92744 | -0.271 |
| ilf3b | 0.417 | zgc:114188 | -0.270 |
| marcksb | 0.417 | apoa1b | -0.250 |
| khdrbs1a | 0.416 | gpx4a | -0.248 |
| smc1al | 0.414 | pnp4b | -0.246 |
| si:ch73-281n10.2 | 0.413 | mat1a | -0.241 |
| anp32a | 0.413 | apoa2 | -0.239 |
| hnrnpaba | 0.412 | agxtb | -0.239 |
| hnrnpabb | 0.412 | nupr1b | -0.235 |
| akap12b | 0.411 | rps17 | -0.235 |
| ncl | 0.406 | apoa4b.1 | -0.232 |
| acin1a | 0.406 | pklr | -0.229 |
| cx43.4 | 0.405 | suclg1 | -0.228 |
| si:ch73-1a9.3 | 0.405 | eno3 | -0.228 |
| hspb1 | 0.405 | grhprb | -0.225 |
| hnrnpa0b | 0.404 | fbp1b | -0.222 |
| si:ch211-152c2.3 | 0.404 | afp4 | -0.220 |
| hnrnpm | 0.403 | aqp12 | -0.218 |
| chaf1a | 0.403 | rbp4 | -0.217 |
| cirbpa | 0.403 | apoc2 | -0.213 |
| ppm1g | 0.401 | aldob | -0.213 |
| snrnp70 | 0.401 | abat | -0.212 |
| hdac1 | 0.400 | fabp10a | -0.212 |
| anp32b | 0.400 | dap | -0.212 |
| hmgb2b | 0.400 | zgc:123103 | -0.212 |
| cbx3a | 0.400 | sod1 | -0.211 |
| lig1 | 0.399 | serpina1 | -0.211 |
| snrpd3l | 0.399 | scp2a | -0.210 |