"SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3a"
ZFIN 
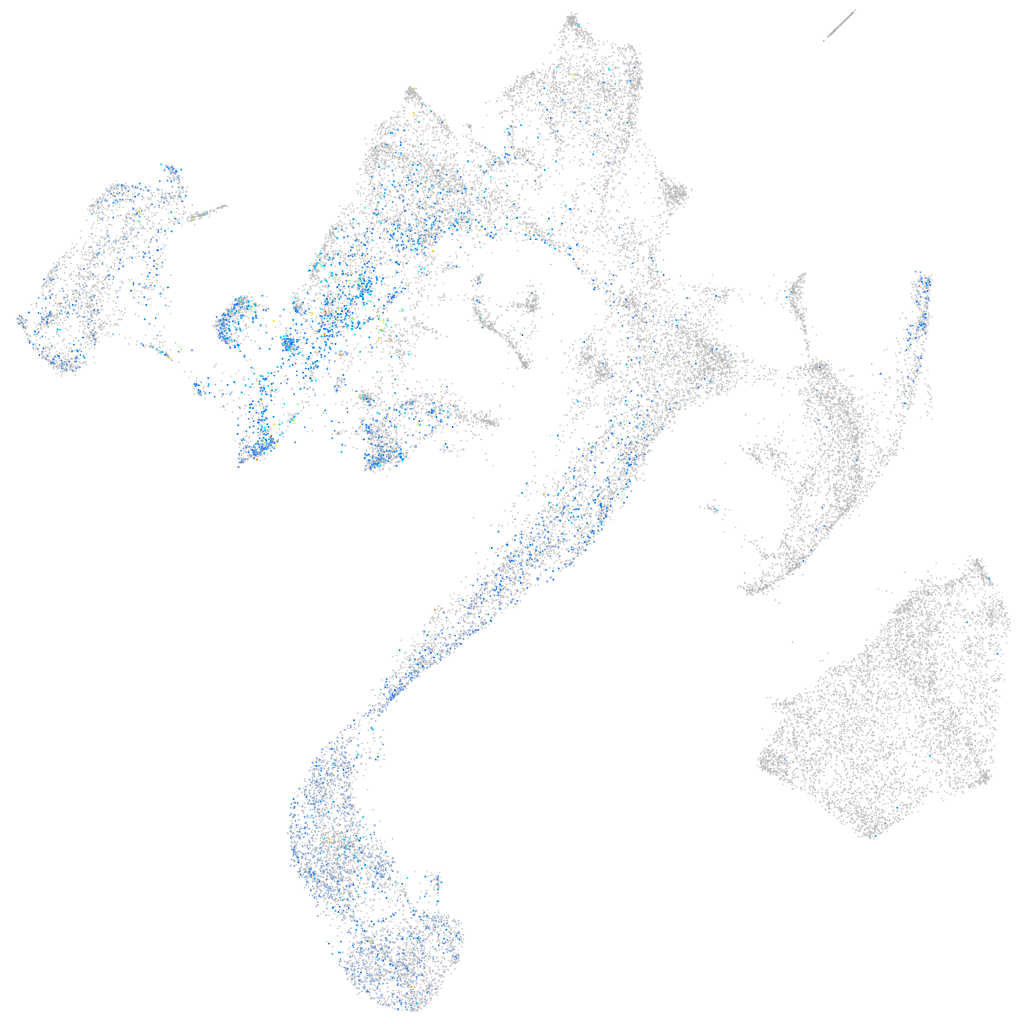
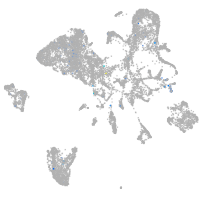
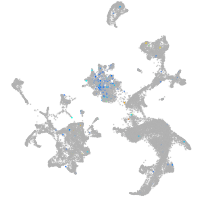
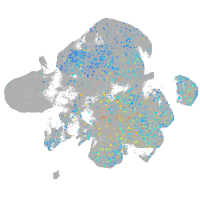
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tmsb | 0.266 | cx43.4 | -0.210 |
| pabpc4 | 0.218 | si:ch73-281n10.2 | -0.195 |
| klhl41b | 0.205 | si:ch211-222l21.1 | -0.189 |
| ttn.2 | 0.204 | apoeb | -0.188 |
| tuba8l3 | 0.202 | si:ch73-1a9.3 | -0.183 |
| actc1b | 0.201 | hmgb2a | -0.182 |
| stac3 | 0.201 | apoc1 | -0.181 |
| gatm | 0.201 | cdx4 | -0.174 |
| casz1 | 0.200 | hes6 | -0.173 |
| tmem38a | 0.199 | ptmab | -0.170 |
| myog | 0.197 | asph | -0.168 |
| desma | 0.195 | hmga1a | -0.167 |
| celf2 | 0.189 | anp32a | -0.166 |
| cav3 | 0.188 | pabpc1a | -0.165 |
| mt-atp6 | 0.186 | fthl27 | -0.165 |
| atp2a1 | 0.183 | nucks1a | -0.163 |
| chrna1 | 0.179 | hsp90ab1 | -0.162 |
| mylpfa | 0.178 | anp32b | -0.161 |
| eef1da | 0.177 | hmgb2b | -0.160 |
| ckmb | 0.175 | h3f3d | -0.157 |
| ckma | 0.174 | hmgn7 | -0.156 |
| COX3 | 0.173 | BX927258.1 | -0.155 |
| eno1a | 0.173 | khdrbs1a | -0.154 |
| acta1a | 0.172 | serpinh1b | -0.154 |
| CABZ01078594.1 | 0.172 | hnrnpaba | -0.153 |
| aldh7a1 | 0.171 | h2afvb | -0.151 |
| phlda2 | 0.170 | hmgn6 | -0.150 |
| ak1 | 0.170 | tuba8l4 | -0.149 |
| cavin4a | 0.170 | acin1a | -0.148 |
| myl1 | 0.169 | seta | -0.148 |
| mt-nd4 | 0.168 | syncrip | -0.148 |
| ahcy | 0.168 | ilf3b | -0.148 |
| mdh2 | 0.167 | sp5l | -0.147 |
| ttn.1 | 0.167 | ube2e1 | -0.146 |
| cox17 | 0.167 | cirbpb | -0.146 |