"SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1b"
ZFIN 
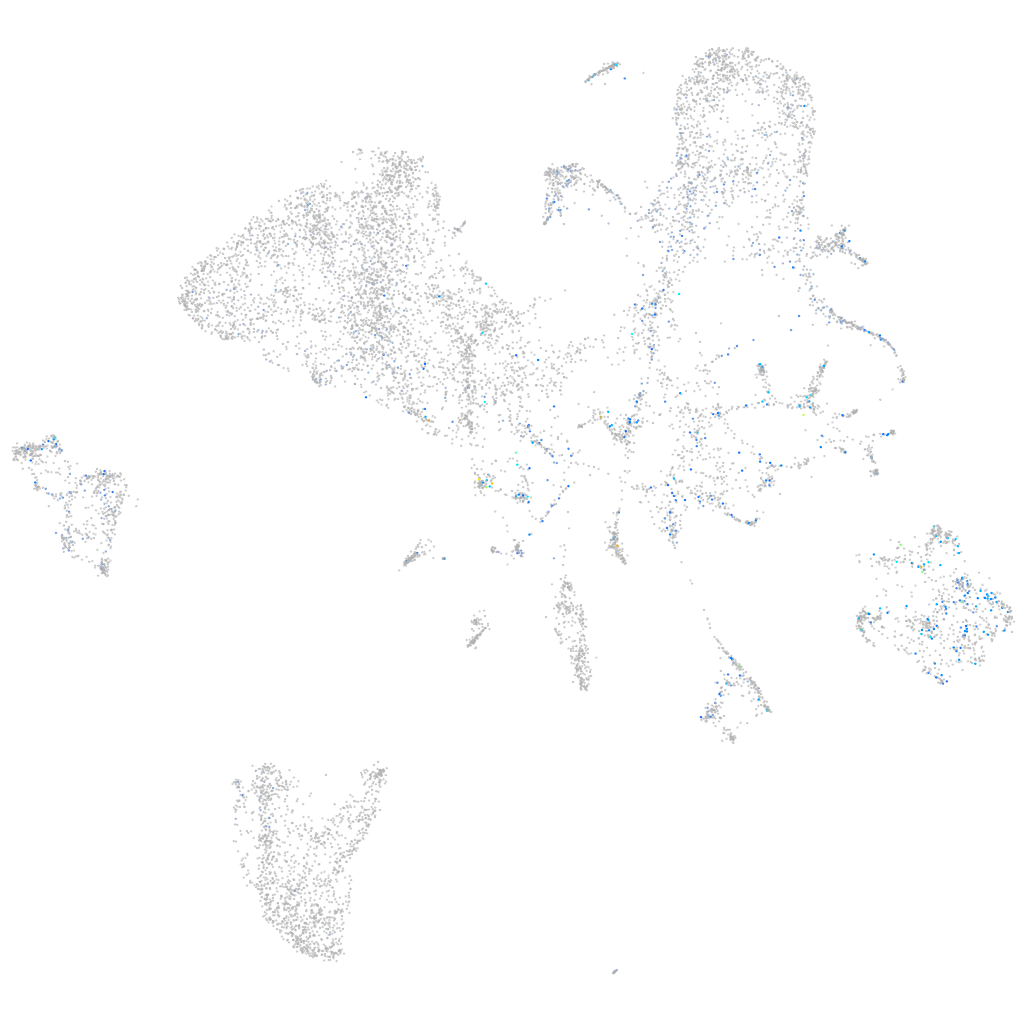
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch73-1a9.3 | 0.174 | gamt | -0.146 |
| nucks1a | 0.173 | gatm | -0.145 |
| si:ch211-222l21.1 | 0.172 | ahcy | -0.138 |
| hnrnpaba | 0.168 | bhmt | -0.132 |
| hnrnpa0b | 0.167 | gapdh | -0.131 |
| syncrip | 0.167 | mat1a | -0.125 |
| hmga1a | 0.167 | zgc:92744 | -0.121 |
| khdrbs1a | 0.167 | aqp12 | -0.120 |
| cirbpa | 0.166 | agxtb | -0.117 |
| hmgn6 | 0.166 | fbp1b | -0.115 |
| seta | 0.166 | pnp4b | -0.115 |
| ilf3b | 0.164 | nupr1b | -0.114 |
| h2afvb | 0.163 | apoa1b | -0.114 |
| hdac1 | 0.162 | apoa4b.1 | -0.112 |
| hmgb2b | 0.162 | apoa2 | -0.111 |
| si:ch211-51e12.7 | 0.159 | afp4 | -0.111 |
| hnrnpub | 0.158 | gpx4a | -0.111 |
| marcksl1b | 0.157 | dap | -0.109 |
| ptmab | 0.157 | rbp4 | -0.105 |
| hmgb2a | 0.157 | apobb.1 | -0.105 |
| setb | 0.157 | grhprb | -0.104 |
| hmgn2 | 0.156 | abat | -0.104 |
| sae1 | 0.156 | apoc2 | -0.103 |
| hnrnpa0a | 0.155 | cx32.3 | -0.103 |
| h3f3d | 0.155 | apom | -0.103 |
| rhoaa | 0.154 | ces2 | -0.102 |
| hdgfl2 | 0.154 | zgc:123103 | -0.101 |
| si:ch73-281n10.2 | 0.153 | fetub | -0.101 |
| acin1a | 0.152 | mgst1.2 | -0.101 |
| top1l | 0.152 | kng1 | -0.101 |
| anp32a | 0.151 | glud1b | -0.101 |
| cbx3a | 0.151 | suclg1 | -0.100 |
| hmgn7 | 0.151 | aldh7a1 | -0.100 |
| rbm4.3 | 0.150 | LOC110437731 | -0.099 |
| hp1bp3 | 0.149 | ttr | -0.098 |