SMAD family member 1
ZFIN 
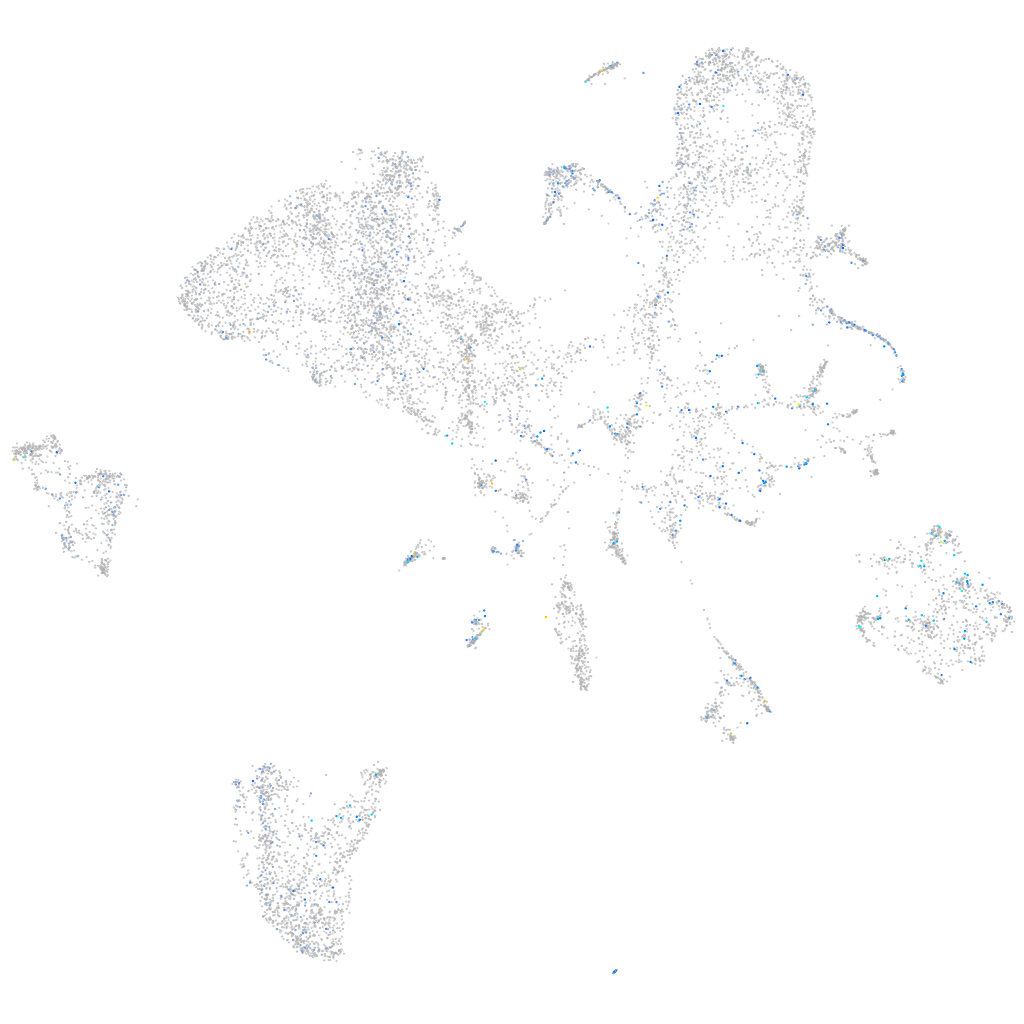
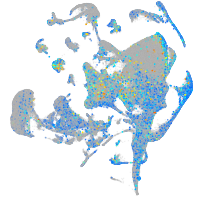
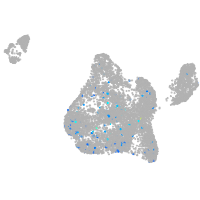
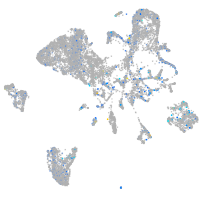
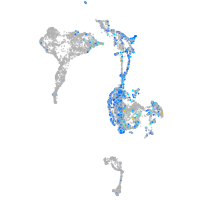
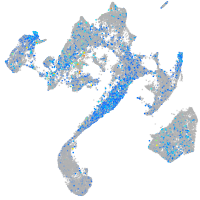
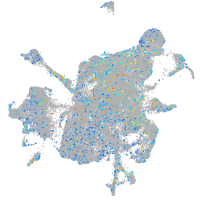
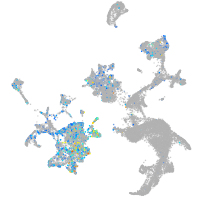
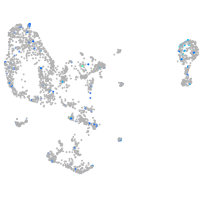
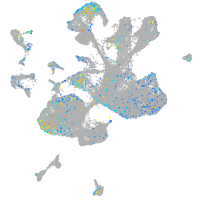
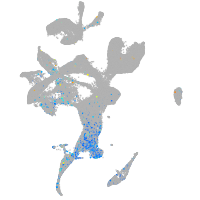
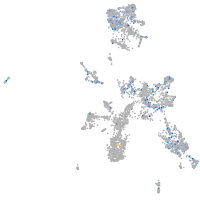
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| psmb11a | 0.134 | gamt | -0.096 |
| kcnj10a | 0.124 | afp4 | -0.092 |
| FO704909.1 | 0.123 | gatm | -0.090 |
| id1 | 0.117 | ahcy | -0.090 |
| npy1r | 0.117 | apoa4b.1 | -0.089 |
| si:dkey-15b23.3 | 0.109 | apoa1b | -0.089 |
| cbln6 | 0.108 | apoc2 | -0.088 |
| mlc1 | 0.107 | gapdh | -0.085 |
| CT025771.1 | 0.105 | gpx4a | -0.078 |
| XLOC-020238 | 0.104 | tfa | -0.078 |
| si:ch211-81a5.8 | 0.104 | mat1a | -0.078 |
| sh3bp2 | 0.103 | fbp1b | -0.078 |
| gprc5c | 0.101 | apobb.1 | -0.077 |
| hyal6 | 0.098 | hao1 | -0.077 |
| si:dkey-7n6.2 | 0.098 | bhmt | -0.076 |
| CU855694.1 | 0.098 | gstt1a | -0.075 |
| cnn2 | 0.096 | slc27a2a | -0.074 |
| pfn1 | 0.096 | apoa2 | -0.074 |
| evx1 | 0.095 | aldob | -0.073 |
| otogl | 0.094 | zgc:123103 | -0.072 |
| chadla | 0.093 | pla2g12b | -0.071 |
| veph1 | 0.092 | suclg2 | -0.071 |
| myct1a | 0.091 | apom | -0.071 |
| capn2b | 0.090 | ftcd | -0.070 |
| elovl1b | 0.090 | fetub | -0.070 |
| spint2 | 0.090 | serpina1l | -0.070 |
| f11r.1 | 0.090 | grhprb | -0.070 |
| gphb5 | 0.090 | sult2st2 | -0.070 |
| hhat | 0.090 | tdo2a | -0.069 |
| slc35f1 | 0.089 | cox7a1 | -0.069 |
| LOC101882326 | 0.089 | rbp2b | -0.069 |
| actb1 | 0.089 | ambp | -0.069 |
| bambia | 0.088 | suclg1 | -0.069 |
| prdm1a | 0.088 | scp2a | -0.068 |
| ca14 | 0.087 | ces2 | -0.068 |