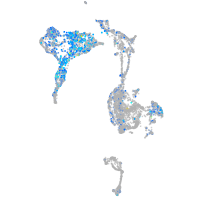
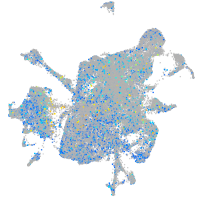
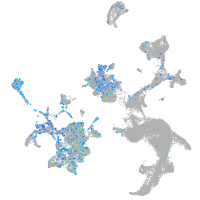
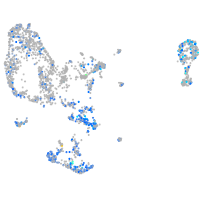
STE20-like kinase b
ZFIN 
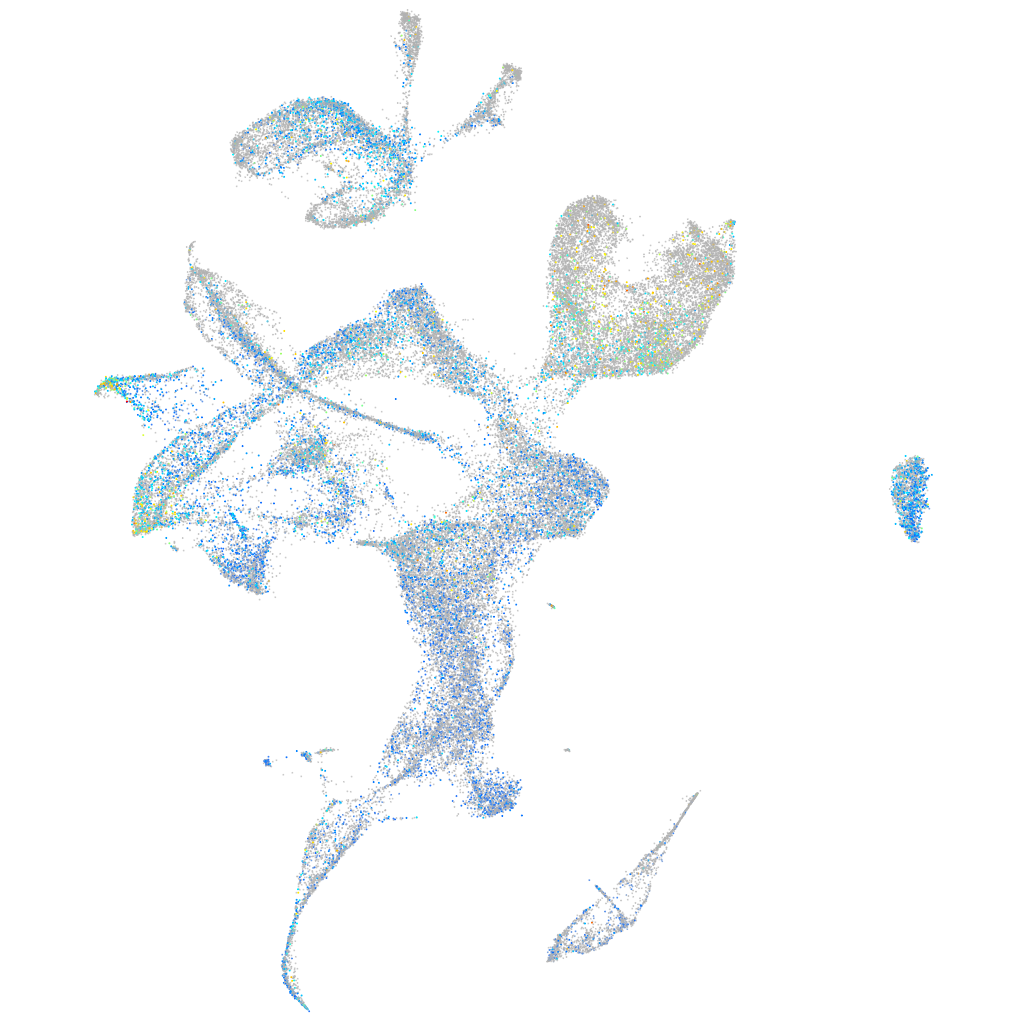
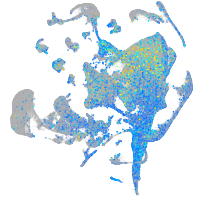
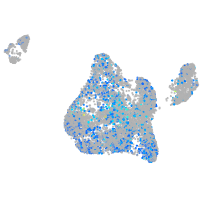
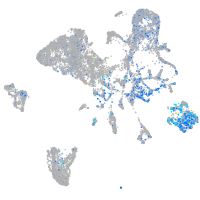
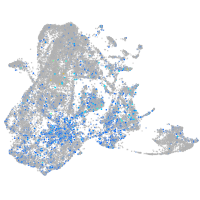
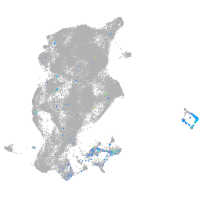
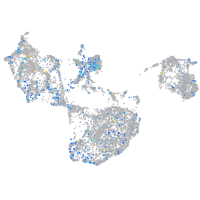
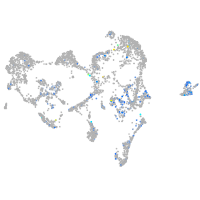
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| saga | 0.158 | si:dkey-16p21.8 | -0.065 |
| pde6b | 0.155 | sncgb | -0.063 |
| pde6ga | 0.154 | olfm1b | -0.058 |
| grk1a | 0.153 | eno1a | -0.053 |
| pde6a | 0.152 | vamp1 | -0.051 |
| sagb | 0.152 | ppiab | -0.050 |
| gngt1 | 0.151 | h3f3a | -0.050 |
| gnat1 | 0.150 | calb2b | -0.049 |
| rom1a | 0.148 | fxyd6l | -0.048 |
| rom1b | 0.147 | si:ch73-256g18.2 | -0.048 |
| pdca | 0.145 | grin1a | -0.048 |
| cnga1b | 0.144 | cabp2a | -0.048 |
| si:ch211-113d22.2 | 0.140 | lin7a | -0.048 |
| cplx4c | 0.140 | nrn1lb | -0.047 |
| guca1b | 0.138 | hmx4 | -0.046 |
| zgc:162144 | 0.135 | atp2b3a | -0.045 |
| rhol | 0.133 | dut | -0.043 |
| cnga1a | 0.132 | actc1b | -0.043 |
| rho | 0.132 | gria3a | -0.043 |
| cngb1a | 0.131 | gnao1b | -0.042 |
| rbp2a | 0.127 | tpi1b | -0.042 |
| CABZ01084566.1 | 0.127 | zgc:165555.3 | -0.042 |
| faimb | 0.127 | calm2b | -0.042 |
| BX004774.2 | 0.125 | si:ch1073-429i10.3.1 | -0.041 |
| gucy2f | 0.118 | kif1aa | -0.041 |
| arl3l2 | 0.117 | si:ch1073-83n3.2 | -0.040 |
| rbp4l | 0.115 | hist1h4l | -0.040 |
| prph2b | 0.114 | zgc:56493 | -0.039 |
| ppdpfa | 0.114 | calm1b | -0.039 |
| pde6h | 0.113 | pvalb1 | -0.039 |
| eno1b | 0.112 | pvalb2 | -0.039 |
| si:ch1073-303d10.1 | 0.111 | ndufa4 | -0.039 |
| pde6ha | 0.110 | FQ323156.1 | -0.039 |
| si:ch211-207j7.2 | 0.109 | rgs16 | -0.038 |
| si:dkey-17e16.15 | 0.109 | stx3a | -0.038 |