solute carrier family 7 member 8a
ZFIN 
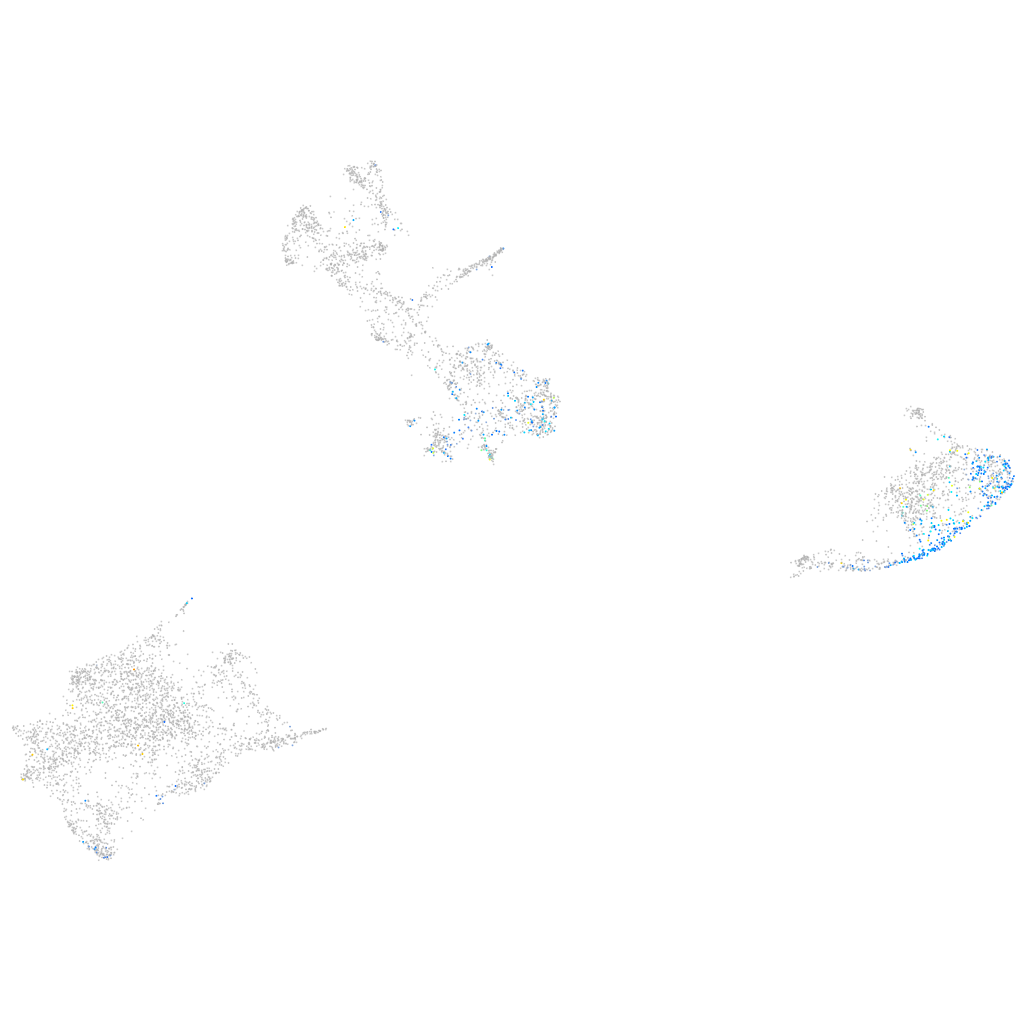
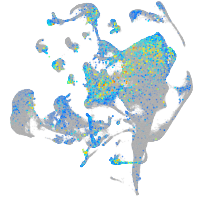
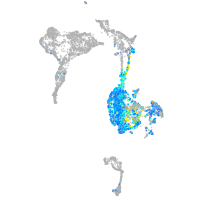
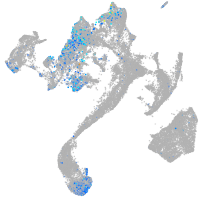
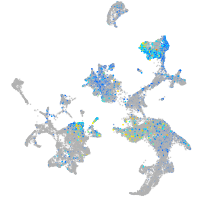
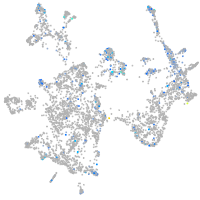
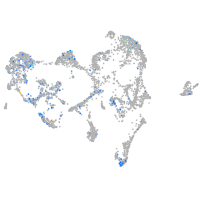
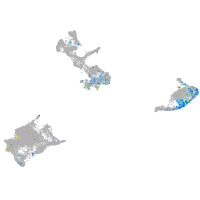
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tyrp1a | 0.352 | paics | -0.214 |
| dct | 0.348 | CABZ01021592.1 | -0.184 |
| tyrp1b | 0.344 | uraha | -0.182 |
| oca2 | 0.342 | atic | -0.164 |
| pmela | 0.341 | impdh1b | -0.163 |
| slc24a5 | 0.340 | si:dkey-251i10.2 | -0.158 |
| aadac | 0.325 | tmem130 | -0.154 |
| kita | 0.324 | slc2a15a | -0.150 |
| zgc:91968 | 0.307 | phyhd1 | -0.144 |
| prkar1b | 0.305 | prps1a | -0.144 |
| slc29a3 | 0.304 | mdh1aa | -0.139 |
| slc7a5 | 0.304 | glulb | -0.135 |
| SPAG9 | 0.300 | ppat | -0.131 |
| tyr | 0.295 | aox5 | -0.128 |
| mtbl | 0.294 | si:ch211-251b21.1 | -0.123 |
| lamp1a | 0.291 | sprb | -0.120 |
| atp6v0a2b | 0.278 | prdx1 | -0.119 |
| slc22a2 | 0.273 | cyb5a | -0.114 |
| si:ch73-389b16.1 | 0.271 | cx30.3 | -0.112 |
| kcnj13 | 0.265 | pax7b | -0.112 |
| slc39a10 | 0.259 | pax7a | -0.110 |
| spra | 0.258 | shmt1 | -0.110 |
| slc24a4a | 0.258 | bscl2l | -0.109 |
| slc30a8 | 0.254 | sult1st1 | -0.107 |
| slc3a2a | 0.254 | gmps | -0.105 |
| gpr61 | 0.253 | TMEM19 | -0.104 |
| pknox1.2 | 0.250 | cax1 | -0.103 |
| slc45a2 | 0.250 | prtfdc1 | -0.103 |
| slc30a1b | 0.248 | bco1 | -0.100 |
| CDK18 | 0.246 | mibp | -0.100 |
| AL935199.1 | 0.246 | prdx5 | -0.099 |
| slc6a15 | 0.245 | apoda.1 | -0.098 |
| nptx1l | 0.244 | rbp4l | -0.095 |
| slc37a2 | 0.241 | tfec | -0.095 |
| hsd20b2 | 0.240 | defbl1 | -0.095 |