solute carrier family 7 member 10b
ZFIN 
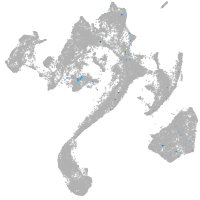
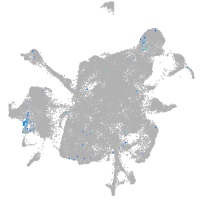
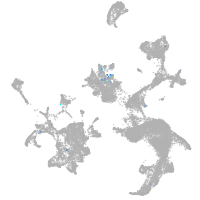
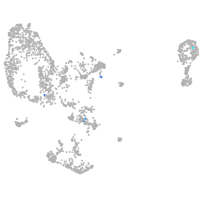
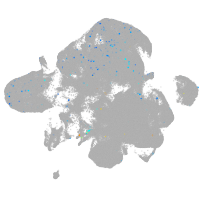
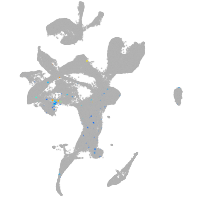
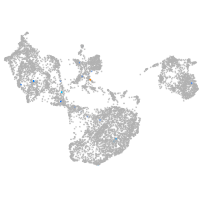
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| slc6a11b | 0.378 | ecscr | -0.015 |
| FO704813.1 | 0.341 | zgc:163057 | -0.015 |
| gpr37l1b | 0.284 | ehd4 | -0.015 |
| slc1a2b | 0.282 | mibp2 | -0.014 |
| hepacama | 0.281 | tie1 | -0.014 |
| mfge8a | 0.272 | lgmn | -0.014 |
| atp1a1b | 0.264 | cdh5 | -0.014 |
| atp1b4 | 0.252 | sox18 | -0.014 |
| slc4a4a | 0.245 | plvapb | -0.014 |
| adra1ab | 0.245 | gp9 | -0.014 |
| hspb15 | 0.243 | mfng | -0.014 |
| cspg5b | 0.240 | clic2 | -0.014 |
| zgc:165461 | 0.232 | fam198b | -0.013 |
| si:ch211-66e2.5 | 0.226 | tcima | -0.013 |
| slc6a1b | 0.225 | she | -0.013 |
| ttll9 | 0.223 | hbbe3 | -0.013 |
| ptgdsb.2 | 0.221 | fli1b | -0.013 |
| LOC103912066 | 0.211 | naga | -0.013 |
| nog2 | 0.209 | clec14a | -0.013 |
| mlc1 | 0.206 | gpr182 | -0.013 |
| bcan | 0.201 | sult2st1 | -0.013 |
| eno1b | 0.192 | cpn1 | -0.013 |
| slc7a10a | 0.192 | glrx | -0.013 |
| slc1a3b | 0.192 | capgb | -0.012 |
| BX005054.1 | 0.191 | si:dkey-27i16.2 | -0.012 |
| gareml | 0.185 | rac2 | -0.012 |
| NPAS3 | 0.180 | tspan36 | -0.012 |
| fjx1 | 0.179 | si:dkeyp-97a10.2 | -0.012 |
| chl1b | 0.179 | tfec | -0.012 |
| grm3 | 0.178 | XLOC-014079 | -0.012 |
| ppap2d | 0.169 | ctsz | -0.012 |
| sept8b | 0.169 | si:ch211-248e11.2 | -0.012 |
| efhd1 | 0.169 | mtmr8 | -0.012 |
| si:ch211-251b21.1 | 0.165 | si:ch211-145b13.6 | -0.012 |
| cldn7a | 0.159 | FERMT3 (1 of many) | -0.012 |