solute carrier family 3 member 1
ZFIN 
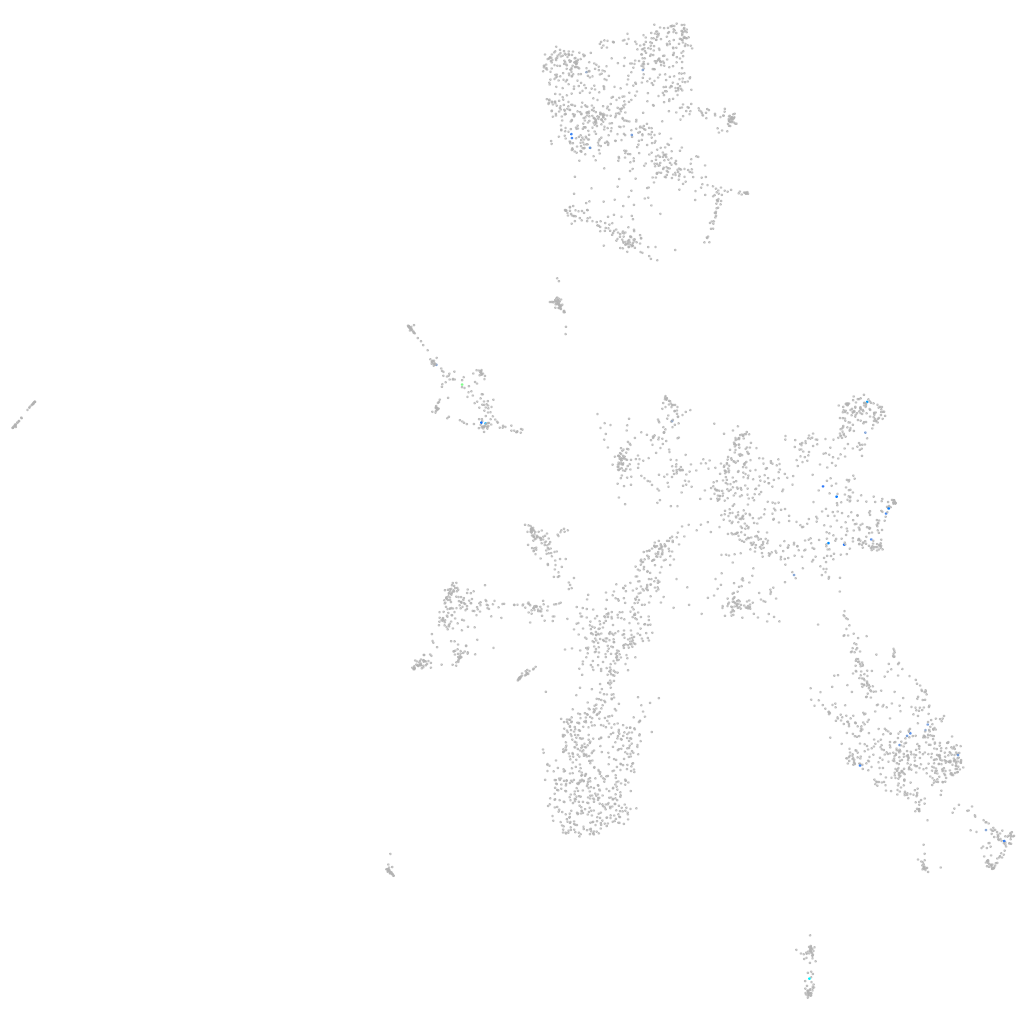
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye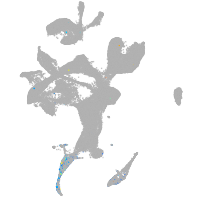 |
taste / olfactory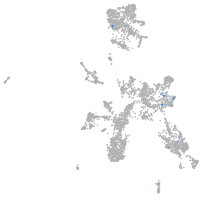 |
otic / lateral line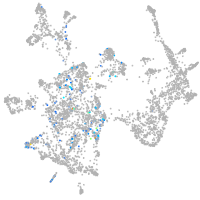 |
epidermis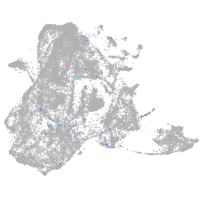 |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CR788303.1 | 0.319 | si:dkey-276j7.1 | -0.048 |
| si:ch1073-220m6.1 | 0.280 | gabarapa | -0.044 |
| si:dkey-26m3.3 | 0.271 | gstp1 | -0.044 |
| cldn15b | 0.263 | srsf9 | -0.042 |
| flt4 | 0.253 | si:ch211-137a8.4 | -0.041 |
| zmp:0000000755 | 0.251 | trappc3 | -0.040 |
| impg1b | 0.242 | atf5a | -0.040 |
| XLOC-044286 | 0.234 | pim3 | -0.039 |
| plekhg2 | 0.220 | gnb1a | -0.038 |
| LOC556776 | 0.219 | bcl11ba | -0.038 |
| kcng1 | 0.213 | insm1b | -0.037 |
| slc27a2a | 0.199 | uqcrq | -0.037 |
| mymk | 0.197 | gnao1b | -0.036 |
| adgre10 | 0.196 | flot2b | -0.036 |
| tomt | 0.194 | atp5f1c | -0.036 |
| ccl39.9 | 0.183 | taf12 | -0.036 |
| samd4a | 0.182 | tmed4 | -0.036 |
| LECT2 (1 of many) | 0.181 | pvalb1 | -0.036 |
| LOC110439792 | 0.174 | znrf1 | -0.036 |
| mipa | 0.172 | pdia6 | -0.035 |
| frem1a | 0.172 | glyr1 | -0.035 |
| FQ311924.2 | 0.171 | rab10 | -0.035 |
| smtlb | 0.169 | txn | -0.034 |
| prox2 | 0.167 | cldnh | -0.034 |
| zgc:114130 | 0.161 | mdh1aa | -0.034 |
| si:ch211-264e16.1 | 0.155 | elavl3 | -0.034 |
| LOC103908767 | 0.151 | fthl31 | -0.034 |
| XLOC-017089 | 0.150 | vamp2 | -0.033 |
| BX546503.1 | 0.149 | stmn1b | -0.033 |
| pcdh1g22 | 0.149 | pvalb2 | -0.033 |
| dusp19a | 0.147 | gdi1 | -0.033 |
| si:dkey-71h2.2 | 0.146 | ap1s3b | -0.033 |
| XLOC-026746 | 0.146 | TSTD1 | -0.033 |
| antxr1c | 0.145 | map1b | -0.033 |
| ppp1r18 | 0.144 | fuca1.1 | -0.033 |