solute carrier family 39 member 9
ZFIN 
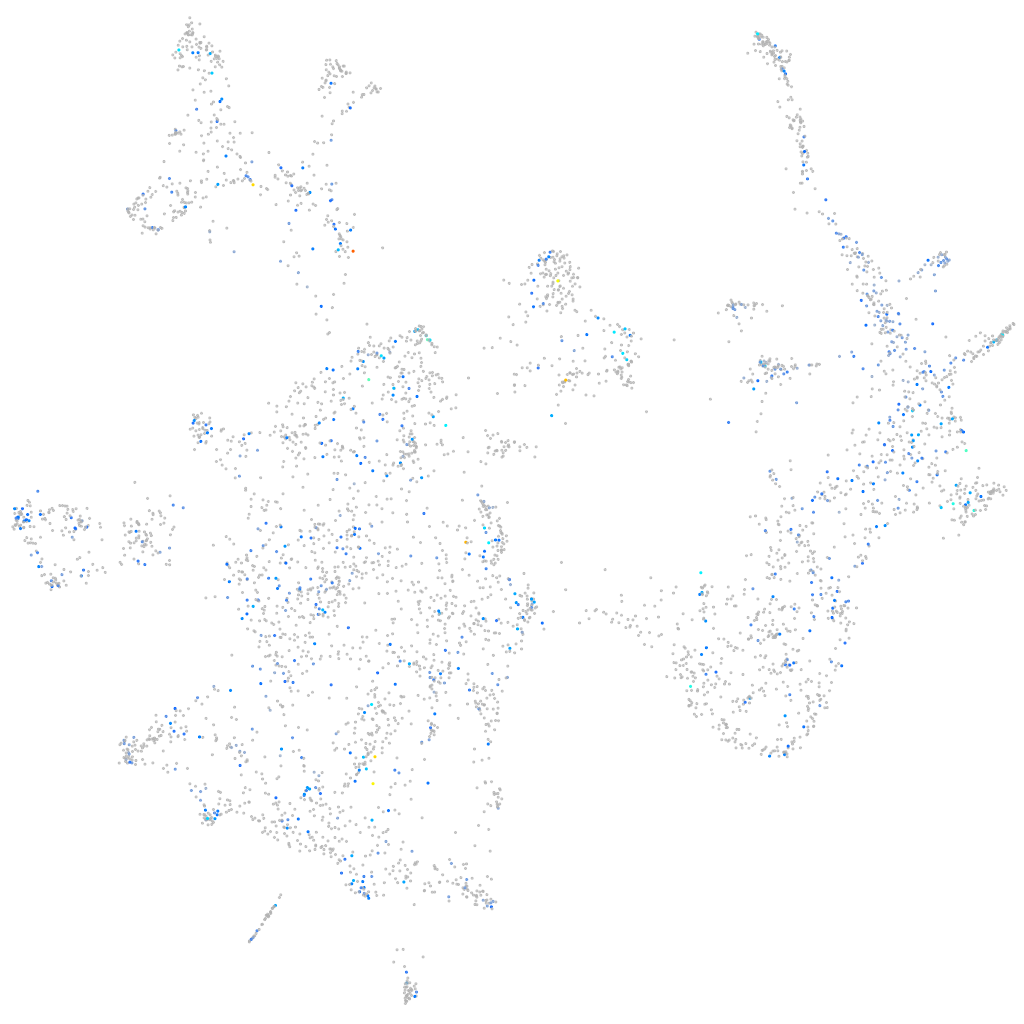
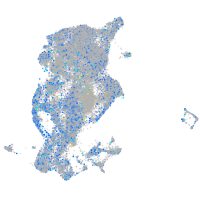
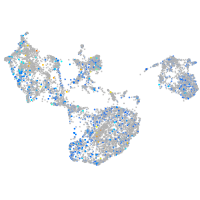
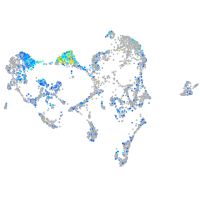
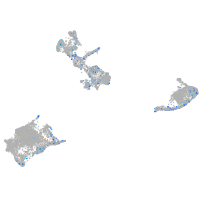
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| prf1.5 | 0.136 | zgc:158463 | -0.070 |
| CU469514.1 | 0.118 | NC-002333.17 | -0.045 |
| CR847530.1 | 0.118 | si:dkey-16p21.8 | -0.042 |
| drgx | 0.116 | s100a1 | -0.042 |
| tmed2 | 0.116 | nmt2 | -0.041 |
| AL935195.1 | 0.114 | smdt1a | -0.041 |
| zgc:174288 | 0.113 | amigo3 | -0.041 |
| ssr3 | 0.112 | atpv0e2 | -0.039 |
| zgc:77197 | 0.109 | hbbe1.1 | -0.038 |
| kcne4 | 0.108 | ctrl | -0.038 |
| ssr4 | 0.106 | serpina1l | -0.038 |
| pdia6 | 0.106 | emb | -0.038 |
| BX548032.1 | 0.103 | cox4i1l | -0.037 |
| ddx5 | 0.102 | tmbim1b | -0.037 |
| XLOC-002129 | 0.101 | atg9a | -0.037 |
| selenof | 0.100 | CR925719.1 | -0.037 |
| pdia3 | 0.100 | gltpb | -0.036 |
| selenom | 0.100 | myo1ca | -0.036 |
| si:ch211-145b13.6 | 0.100 | COX3 | -0.036 |
| calr3b | 0.099 | tekt3 | -0.036 |
| calr | 0.099 | calm1b | -0.036 |
| LOC799656 | 0.099 | CU138547.1 | -0.036 |
| tmed9 | 0.098 | KCNIP4 | -0.035 |
| sdf2l1 | 0.098 | si:dkeyp-75h12.2 | -0.035 |
| cntnap1 | 0.097 | prss59.1 | -0.035 |
| si:dkeyp-80c12.7 | 0.096 | myhz1.2 | -0.034 |
| CR293531.2 | 0.095 | gpx2 | -0.034 |
| hyou1 | 0.095 | prss59.2 | -0.034 |
| hoxa4a | 0.095 | apoa2 | -0.034 |
| dnajb11 | 0.095 | rab4b | -0.034 |
| CU013548.1 | 0.095 | pcloa | -0.034 |
| aste1a | 0.094 | fabp1b.1 | -0.034 |
| mydgf | 0.094 | si:ch211-258f14.2 | -0.034 |
| si:ch211-210b2.3 | 0.094 | tfa | -0.033 |
| cav3 | 0.094 | matn1 | -0.033 |