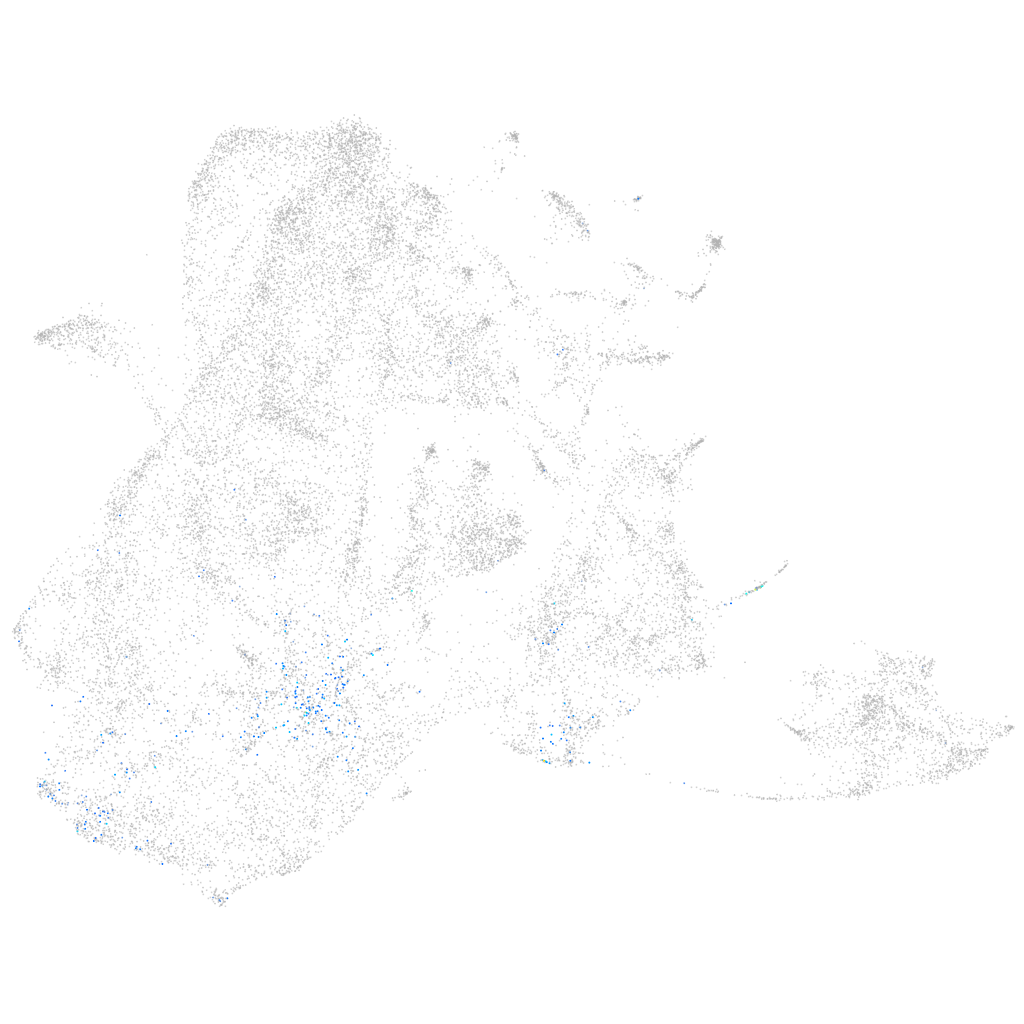
solute carrier family 39 member 4
ZFIN 
Other cell groups


all cells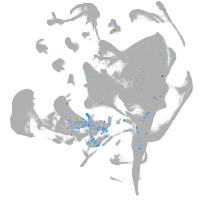 |
blastomeres |
PGCs |
endoderm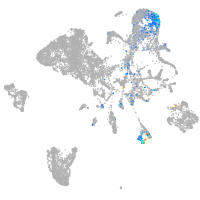 |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye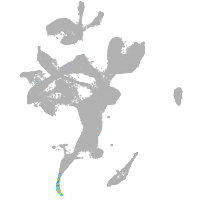 |
taste / olfactory |
otic / lateral line |
epidermis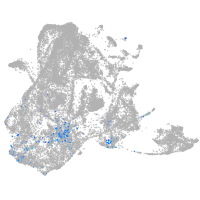 |
periderm |
fin |
ionocytes / mucous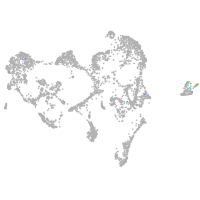 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| six1b | 0.188 | tmsb1 | -0.185 |
| apela | 0.180 | krt4 | -0.146 |
| foxi1 | 0.177 | s100a10b | -0.146 |
| dclk3 | 0.168 | pfn1 | -0.140 |
| eya1 | 0.163 | cfl1l | -0.140 |
| si:ch211-67e16.11 | 0.163 | cotl1 | -0.126 |
| zgc:194210 | 0.162 | capns1a | -0.124 |
| cx43.4 | 0.156 | zgc:162730 | -0.120 |
| insm1b | 0.154 | cyt1 | -0.118 |
| si:ch211-152c2.3 | 0.154 | aqp3a | -0.118 |
| her15.1 | 0.152 | anxa1a | -0.118 |
| myl10 | 0.147 | krt91 | -0.115 |
| six4b | 0.147 | col1a1b | -0.114 |
| zbtb16a | 0.147 | gapdhs | -0.112 |
| akirin1 | 0.143 | tmsb4x | -0.111 |
| tspan7 | 0.138 | cldn1 | -0.109 |
| khdrbs1a | 0.137 | eef1da | -0.108 |
| tmeff1b | 0.135 | cebpd | -0.107 |
| itga6a | 0.134 | col1a2 | -0.107 |
| cdkn1ca | 0.132 | col1a1a | -0.106 |
| greb1l | 0.132 | krtt1c19e | -0.106 |
| hdac1 | 0.130 | krt5 | -0.103 |
| snrpe | 0.129 | rbp4 | -0.102 |
| marcksb | 0.128 | pycard | -0.102 |
| cited4b | 0.128 | wu:fb18f06 | -0.100 |
| apex1 | 0.128 | anxa2a | -0.098 |
| sumo3b | 0.128 | postnb | -0.098 |
| si:dkey-42i9.4 | 0.128 | si:dkey-33c14.3 | -0.098 |
| hmga1a | 0.127 | spaca4l | -0.095 |
| ilf2 | 0.126 | cldni | -0.094 |
| hnrnpabb | 0.126 | sparc | -0.093 |
| hnrnpaba | 0.126 | rplp2 | -0.093 |
| lin28a | 0.126 | rac2 | -0.093 |
| hnrnpa0l | 0.125 | col11a1a | -0.092 |
| smarce1 | 0.125 | col4a5 | -0.092 |