solute carrier family 39 member 1
ZFIN 
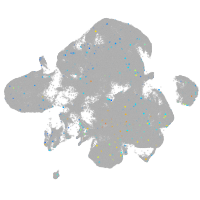
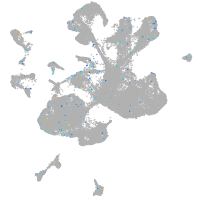
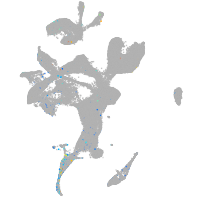
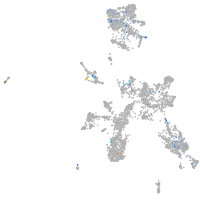
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| kita | 0.356 | CABZ01021592.1 | -0.181 |
| tyrp1b | 0.338 | paics | -0.162 |
| pmela | 0.338 | uraha | -0.148 |
| oca2 | 0.337 | si:dkey-251i10.2 | -0.141 |
| dct | 0.335 | tmem130 | -0.127 |
| tyrp1a | 0.332 | mdh1aa | -0.122 |
| slc24a5 | 0.329 | atic | -0.108 |
| slc22a2 | 0.322 | phyhd1 | -0.096 |
| SPAG9 | 0.322 | cyb5a | -0.094 |
| slc7a5 | 0.302 | impdh1b | -0.093 |
| zgc:91968 | 0.302 | si:ch211-251b21.1 | -0.090 |
| slc39a10 | 0.301 | si:ch211-222l21.1 | -0.090 |
| tyr | 0.301 | mdkb | -0.087 |
| lamp1a | 0.294 | slc2a15a | -0.087 |
| si:ch73-389b16.1 | 0.294 | aox5 | -0.085 |
| prkar1b | 0.292 | prps1a | -0.084 |
| slc45a2 | 0.284 | glulb | -0.083 |
| mtbl | 0.284 | si:ch73-1a9.3 | -0.082 |
| aadac | 0.275 | sult1st1 | -0.080 |
| zdhhc2 | 0.266 | bco1 | -0.077 |
| megf10 | 0.265 | hmgn2 | -0.076 |
| tspan10 | 0.264 | prdx1 | -0.076 |
| kcnj13 | 0.262 | ptmaa | -0.076 |
| slc3a2a | 0.261 | hbae3 | -0.075 |
| pknox1.2 | 0.261 | mdka | -0.074 |
| mlpha | 0.259 | krt18b | -0.071 |
| CDK18 | 0.254 | bscl2l | -0.071 |
| hsd20b2 | 0.253 | zgc:153031 | -0.071 |
| slc37a2 | 0.251 | defbl1 | -0.070 |
| slc6a15 | 0.251 | rbp4l | -0.069 |
| tspan36 | 0.251 | hbbe1.1 | -0.069 |
| atp6v0a2b | 0.250 | aldob | -0.069 |
| slc29a3 | 0.249 | oacyl | -0.068 |
| spra | 0.249 | hbbe1.3 | -0.067 |
| tfap2e | 0.249 | apoda.1 | -0.067 |