"solute carrier family 26 member 3, tandem duplicate 2"
ZFIN 
Other cell groups


all cells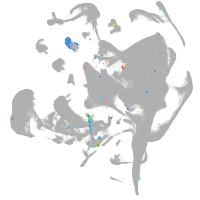 |
blastomeres |
PGCs |
endoderm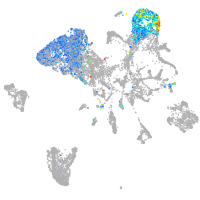 |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle 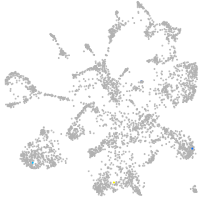 |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory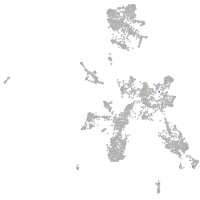 |
otic / lateral line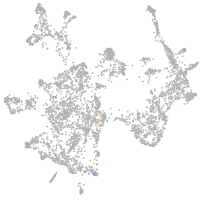 |
epidermis |
periderm |
fin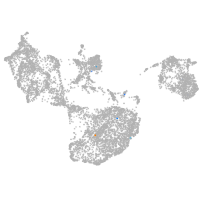 |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| zgc:158432 | 0.668 | mt-co1 | -0.041 |
| nr5a5 | 0.648 | pabpc1a | -0.030 |
| XLOC-041002 | 0.625 | cox8a | -0.027 |
| myct1a | 0.565 | mt-co2 | -0.026 |
| zgc:153968 | 0.512 | atp5mf | -0.026 |
| CABZ01001434.1 | 0.496 | ube2v2 | -0.026 |
| sult1st2 | 0.490 | gng5 | -0.024 |
| ace2 | 0.482 | calm3b | -0.024 |
| zgc:77748 | 0.378 | tomm7 | -0.023 |
| fybb | 0.360 | map2k6 | -0.023 |
| myo7bb | 0.343 | ola1 | -0.023 |
| LOC101886275 | 0.328 | srsf9 | -0.023 |
| si:ch211-220f21.2 | 0.327 | arpc3 | -0.023 |
| LOC110438492 | 0.327 | CABZ01080702.1 | -0.022 |
| slc16a4 | 0.327 | si:dkey-28b4.7 | -0.022 |
| dgat2 | 0.321 | atp5pd | -0.022 |
| cers3a | 0.320 | vdac1 | -0.022 |
| nkx1.2lb | 0.317 | cdc42l | -0.022 |
| ins | 0.311 | selenot2 | -0.022 |
| bglap | 0.302 | dedd1 | -0.022 |
| serpinc1 | 0.298 | igf2bp3 | -0.022 |
| abo | 0.290 | ndufb3 | -0.022 |
| CR812836.1 | 0.270 | dad1 | -0.022 |
| chia.2 | 0.267 | atrx | -0.021 |
| myoz1a | 0.264 | abracl | -0.021 |
| cldn15a | 0.261 | calm1a | -0.021 |
| si:ch211-107e6.6 | 0.254 | atp5f1c | -0.021 |
| hoxa9b | 0.249 | spcs2 | -0.021 |
| tmem86b | 0.247 | calm2a | -0.021 |
| CABZ01060936.1 | 0.247 | rab2a | -0.021 |
| parp9 | 0.243 | erh | -0.021 |
| rsad1 | 0.240 | ap2m1a | -0.021 |
| fga | 0.239 | dcun1d5 | -0.020 |
| six5 | 0.231 | zfp36l1a | -0.020 |
| ca4b | 0.222 | stx12 | -0.020 |