solute carrier family 26 member 1
ZFIN 
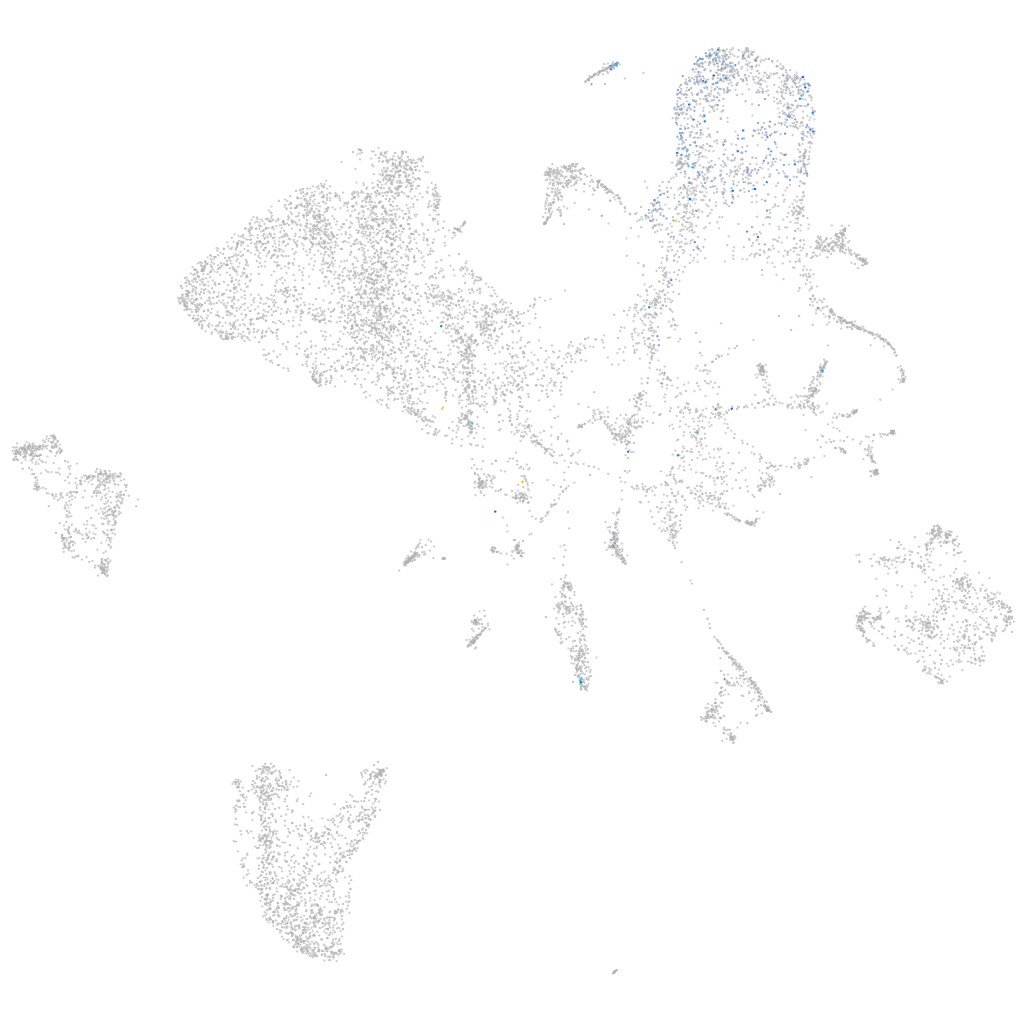
Other cell groups


Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| aqp8a.2 | 0.303 | fabp3 | -0.154 |
| ace2 | 0.300 | aqp12 | -0.125 |
| slc6a19a.2 | 0.295 | bhmt | -0.116 |
| cpo | 0.288 | agxtb | -0.107 |
| mogat2 | 0.286 | apoc1 | -0.106 |
| cdh17 | 0.283 | tfa | -0.102 |
| cd36 | 0.281 | rbp2b | -0.100 |
| ldha | 0.279 | fabp10a | -0.100 |
| vwa11 | 0.278 | ttc36 | -0.100 |
| mbl2 | 0.277 | vtnb | -0.100 |
| atp1a1a.4 | 0.276 | wu:fj16a03 | -0.100 |
| slc15a1b | 0.275 | ambp | -0.099 |
| vil1 | 0.274 | hpda | -0.099 |
| chia.2 | 0.274 | cpn1 | -0.099 |
| rtn4b | 0.273 | apoa2 | -0.098 |
| zgc:158846 | 0.270 | zgc:123103 | -0.098 |
| si:ch211-133l5.7 | 0.269 | fgg | -0.098 |
| mansc1 | 0.269 | uox | -0.098 |
| zgc:153968 | 0.269 | fgb | -0.097 |
| wu:fa56d06 | 0.269 | serpina1 | -0.097 |
| zgc:103681 | 0.267 | apom | -0.096 |
| ckmt1 | 0.266 | kng1 | -0.096 |
| acsl5 | 0.265 | fga | -0.096 |
| fabp1b.1 | 0.265 | ttr | -0.096 |
| si:ch211-127i16.2 | 0.262 | rbp4 | -0.096 |
| ada | 0.262 | cfhl4 | -0.095 |
| meltf | 0.260 | c9 | -0.095 |
| aoc1 | 0.260 | ces2 | -0.095 |
| ndufa4 | 0.258 | gc | -0.095 |
| plac8.1 | 0.258 | igfbp1a | -0.094 |
| lta4h | 0.257 | serpina1l | -0.094 |
| pdzk1 | 0.257 | pygl | -0.094 |
| slc5a1 | 0.255 | zgc:112265 | -0.094 |
| sdsl | 0.254 | fxyd1 | -0.093 |
| AL831745.1 | 0.254 | f2 | -0.092 |