solute carrier family 25 member 36a
ZFIN 
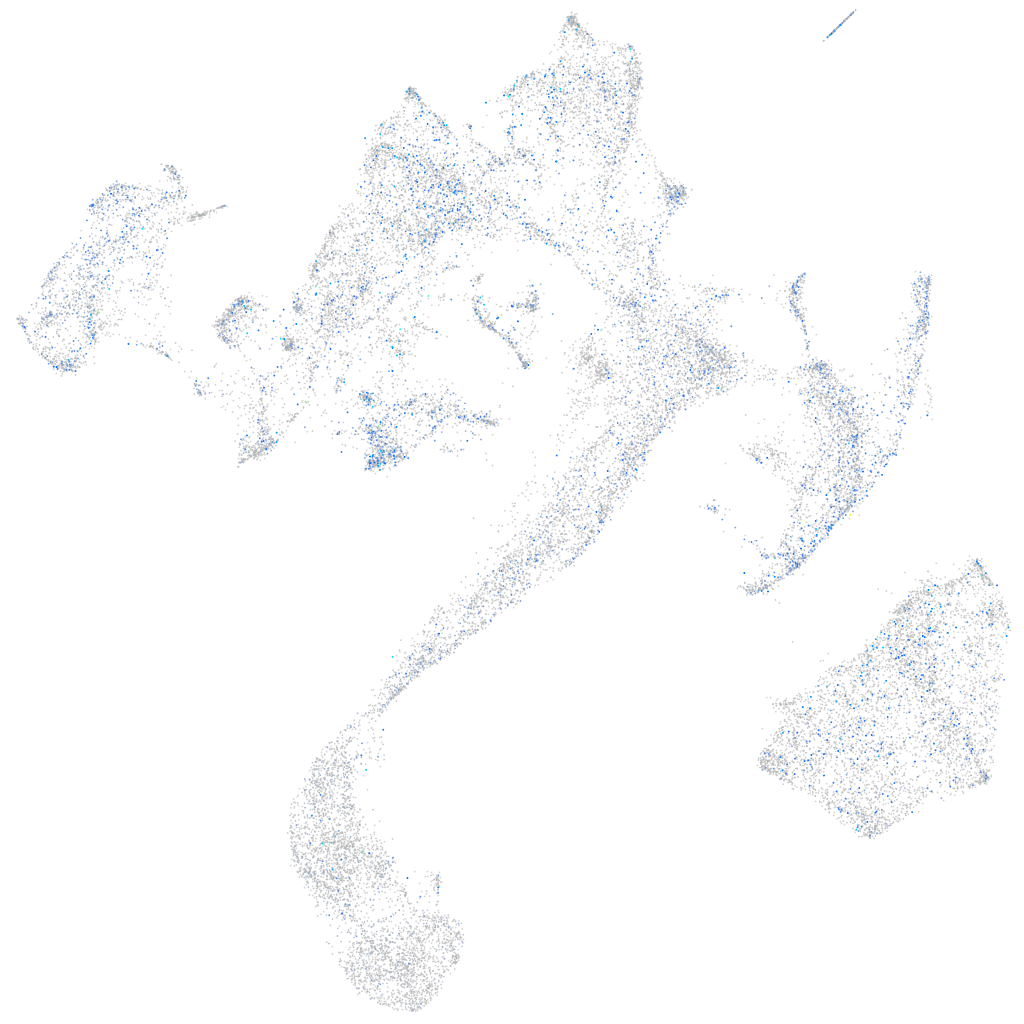
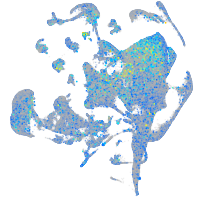
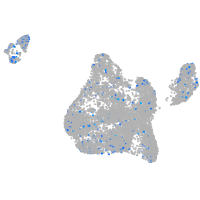
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CR383676.1 | 0.093 | mylpfb | -0.065 |
| ptmaa | 0.073 | myoz1b | -0.064 |
| NC-002333.17 | 0.071 | myl1 | -0.063 |
| gabarapb | 0.069 | tnni2a.4 | -0.062 |
| atp6v1e1b | 0.068 | slc25a4 | -0.062 |
| mt-cyb | 0.068 | mylz3 | -0.061 |
| kdm6bb | 0.067 | myom2a | -0.060 |
| rnasekb | 0.066 | atp2a1l | -0.060 |
| si:dkey-261h17.1 | 0.066 | CABZ01061524.1 | -0.059 |
| stmn1b | 0.066 | casq1a | -0.059 |
| COX3 | 0.065 | tnnt3b | -0.058 |
| tuba1c | 0.065 | pvalb2 | -0.058 |
| myh7bb | 0.064 | ryr3 | -0.056 |
| mt-co2 | 0.063 | hsc70 | -0.054 |
| rarga | 0.063 | pvalb1 | -0.054 |
| calm1a | 0.063 | myhz1.1 | -0.054 |
| tpm3 | 0.063 | mylpfa | -0.052 |
| wu:fb55g09 | 0.063 | tpma | -0.052 |
| fam117ba | 0.062 | myhz1.2 | -0.052 |
| gpm6aa | 0.062 | myhz1.3 | -0.050 |
| myoz2b | 0.062 | ampd1 | -0.049 |
| mxd1 | 0.062 | si:ch211-152c2.3 | -0.048 |
| cdh2 | 0.062 | myoz1a | -0.048 |
| bsg | 0.061 | gyg1b | -0.047 |
| si:ch211-196l7.4 | 0.061 | rgcc | -0.045 |
| si:dkey-56f14.7 | 0.061 | igfn1.3 | -0.044 |
| marcksl1a | 0.061 | si:ch73-367p23.2 | -0.043 |
| alcama | 0.060 | si:ch211-255p10.3 | -0.042 |
| elavl3 | 0.060 | myoz2a | -0.041 |
| camk2n1a | 0.060 | tnnt3a | -0.041 |
| h3f3c | 0.060 | gatm | -0.040 |
| sfrp2 | 0.060 | hhatla | -0.040 |
| zgc:101840 | 0.059 | myhz2 | -0.039 |
| mylk3 | 0.059 | eef2l2 | -0.039 |
| ckbb | 0.059 | pvalb3 | -0.039 |