solute carrier family 25 member 33
ZFIN 
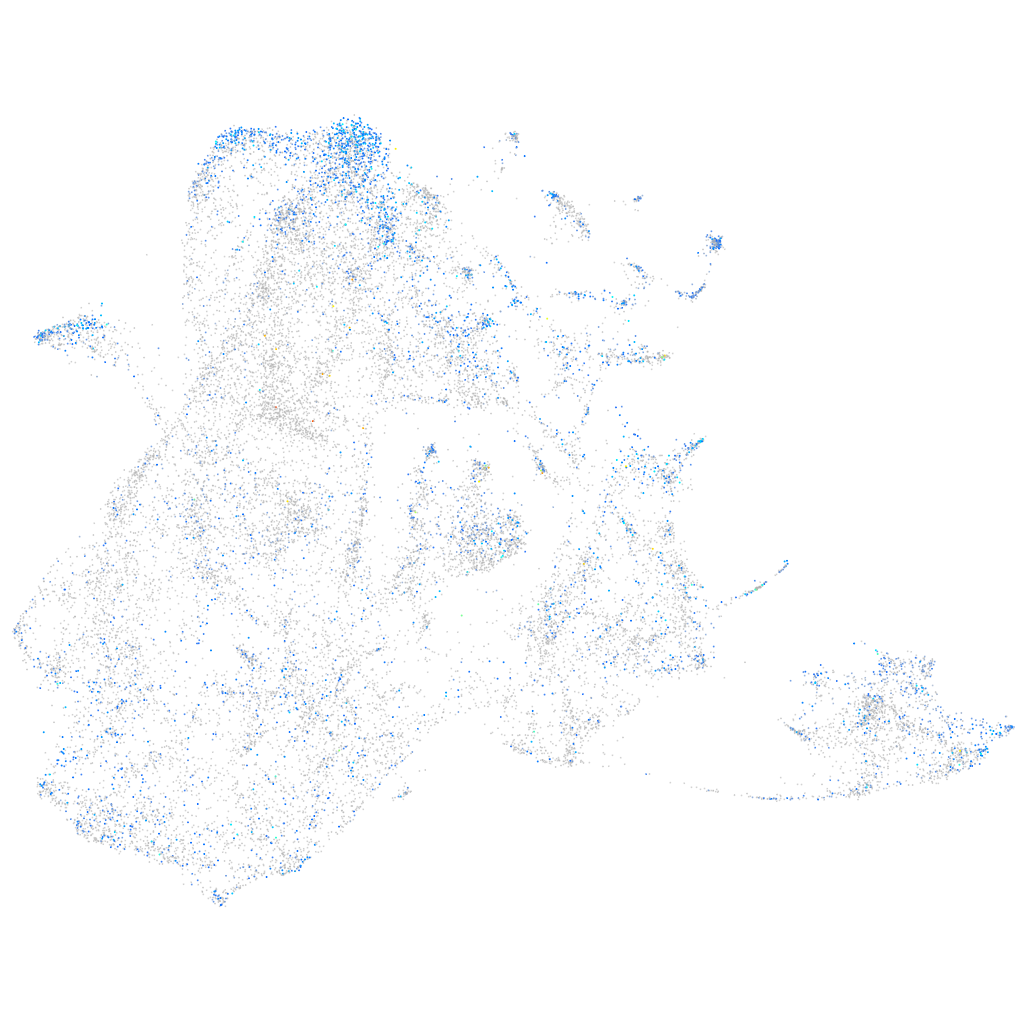
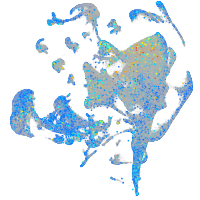
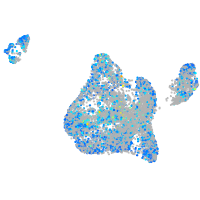
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tdh | 0.233 | NC-002333.4 | -0.130 |
| prelid3b | 0.223 | si:ch73-1a9.3 | -0.129 |
| pnp5a | 0.213 | si:ch211-222l21.1 | -0.113 |
| cdkn1a | 0.205 | si:ch73-46j18.5 | -0.109 |
| ppifb | 0.191 | NC-002333.17 | -0.107 |
| ddx21 | 0.190 | ptmab | -0.102 |
| gtpbp4 | 0.186 | hmgn6 | -0.100 |
| ahcy | 0.182 | hmga1a | -0.091 |
| dusp2 | 0.180 | ost4 | -0.087 |
| rsl24d1 | 0.176 | CABZ01072614.1 | -0.086 |
| eif4ebp3 | 0.176 | si:ch73-281n10.2 | -0.085 |
| cebpb | 0.172 | serpinh1b | -0.085 |
| higd1a | 0.169 | fkbp9 | -0.084 |
| epgn | 0.169 | hbbe1.3 | -0.084 |
| mcl1a | 0.168 | hbae3 | -0.080 |
| zgc:77439 | 0.161 | LOC110439372 | -0.080 |
| ucp2 | 0.159 | hmgb3a | -0.080 |
| nop53 | 0.158 | nucks1a | -0.079 |
| eef1g | 0.155 | tmsb4x | -0.079 |
| tob1b | 0.155 | ppib | -0.078 |
| pno1 | 0.154 | gsnb | -0.076 |
| rsl1d1 | 0.153 | mt-co1 | -0.076 |
| eef1da | 0.150 | col28a2a | -0.075 |
| romo1 | 0.149 | lrata | -0.074 |
| midn | 0.149 | scinla | -0.073 |
| bccip | 0.146 | fkbp1aa | -0.073 |
| arg2 | 0.145 | asph | -0.072 |
| dars | 0.145 | hbae1.1 | -0.070 |
| mylipb | 0.144 | marcksl1b | -0.070 |
| btf3 | 0.144 | h2afvb | -0.069 |
| chac1 | 0.142 | hp1bp3 | -0.069 |
| sat1a.2 | 0.141 | sub1a | -0.068 |
| cebpd | 0.139 | alox12 | -0.067 |
| eef1b2 | 0.139 | p4ha1b | -0.066 |
| ddx5 | 0.139 | zgc:158463 | -0.066 |