solute carrier family 25 member 25b
ZFIN 
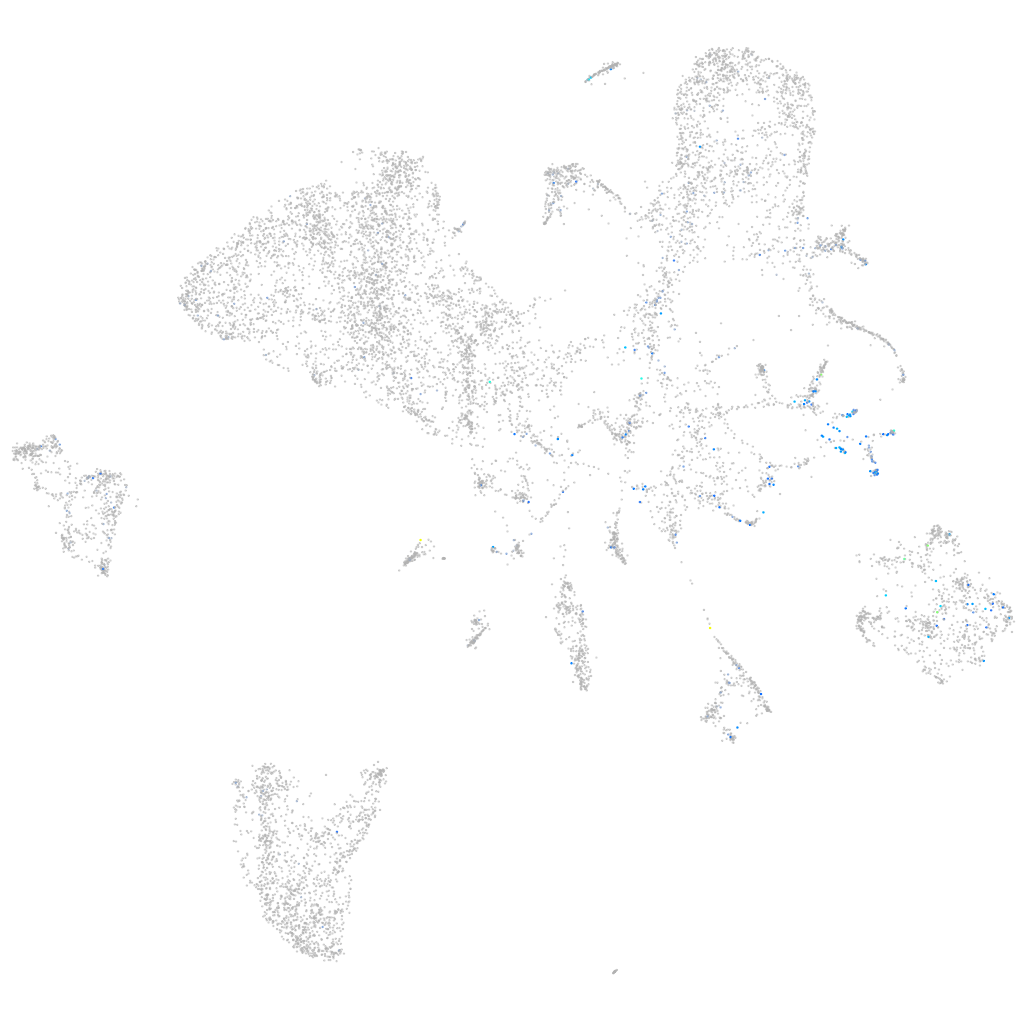
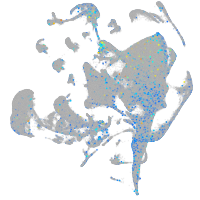
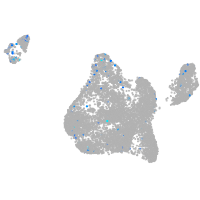
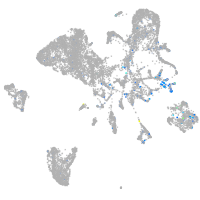
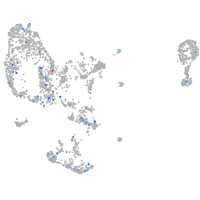
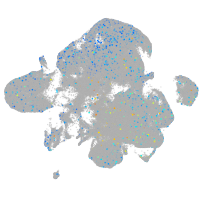
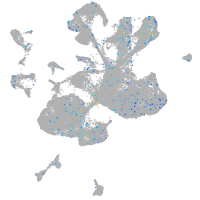
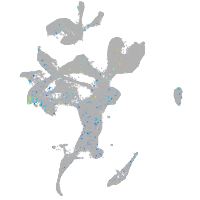
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scgn | 0.257 | gapdh | -0.122 |
| pax6b | 0.253 | ahcy | -0.117 |
| gcgb | 0.246 | gamt | -0.117 |
| ndufa4l2a | 0.245 | gatm | -0.110 |
| slc30a2 | 0.245 | aldob | -0.106 |
| pcsk1nl | 0.242 | bhmt | -0.102 |
| isl1 | 0.241 | mat1a | -0.099 |
| si:dkey-153k10.9 | 0.240 | nupr1b | -0.098 |
| kcnj11 | 0.237 | apoa1b | -0.096 |
| ins | 0.234 | apoa4b.1 | -0.096 |
| neurod1 | 0.229 | fbp1b | -0.094 |
| scg3 | 0.227 | aldh6a1 | -0.093 |
| abhd15a | 0.217 | apoc2 | -0.092 |
| rprmb | 0.216 | glud1b | -0.092 |
| pcsk2 | 0.207 | agxtb | -0.091 |
| ptn | 0.203 | afp4 | -0.091 |
| insm1a | 0.203 | aldh7a1 | -0.088 |
| SLC5A10 | 0.201 | apoa2 | -0.087 |
| pcsk1 | 0.199 | eno3 | -0.087 |
| LOC100537384 | 0.198 | cx32.3 | -0.087 |
| mir7a-1 | 0.197 | mgst1.2 | -0.087 |
| syt1a | 0.193 | scp2a | -0.086 |
| kcnk9 | 0.192 | ckba | -0.086 |
| scg2b | 0.191 | gpx4a | -0.085 |
| renbp | 0.188 | sod1 | -0.083 |
| adm2a | 0.187 | gnmt | -0.083 |
| gpr22a | 0.187 | grhprb | -0.082 |
| elfn1a | 0.187 | aqp12 | -0.081 |
| tspan7b | 0.184 | apobb.1 | -0.081 |
| myt1b | 0.183 | pnp4b | -0.081 |
| plppr5b | 0.182 | dap | -0.081 |
| vat1 | 0.181 | gstt1a | -0.081 |
| dkk3b | 0.181 | suclg1 | -0.080 |
| rims2a | 0.180 | fabp10a | -0.080 |
| scg2a | 0.179 | agxta | -0.080 |