solute carrier family 25 member 24
ZFIN 
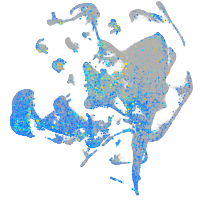
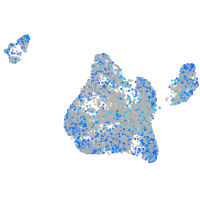
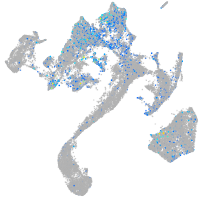
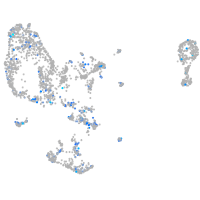
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pnocb | 0.150 | pvalb1 | -0.065 |
| lmod2a | 0.150 | suv39h1b | -0.060 |
| ftr82 | 0.145 | slc29a1a | -0.055 |
| akap12b | 0.141 | eva1bb | -0.054 |
| cavin2b | 0.140 | zgc:110216 | -0.053 |
| krt8 | 0.138 | pvalb2 | -0.052 |
| ppfibp1a | 0.137 | tfpia | -0.052 |
| myh10 | 0.132 | ahcy | -0.051 |
| podxl | 0.128 | stmn1a | -0.051 |
| krt18a.1 | 0.127 | CELA1 (1 of many) | -0.050 |
| ahnak | 0.125 | atp1b4 | -0.050 |
| jam2b | 0.122 | apoa2 | -0.050 |
| myl7 | 0.121 | igfbp7 | -0.049 |
| ywhaz | 0.120 | nop56 | -0.049 |
| CABZ01075068.1 | 0.119 | prss59.2 | -0.049 |
| bnc2 | 0.119 | apoa1b | -0.049 |
| phactr4a | 0.119 | inka1a | -0.049 |
| tnni1b | 0.118 | pcna | -0.048 |
| cav1 | 0.118 | rplp1 | -0.048 |
| krt94 | 0.117 | pcdh18a | -0.047 |
| slc29a1b | 0.117 | si:ch211-251b21.1 | -0.047 |
| cotl1 | 0.116 | per3 | -0.047 |
| angpt1 | 0.113 | prss59.1 | -0.047 |
| si:ch211-198m17.1 | 0.113 | si:ch211-195b13.1 | -0.046 |
| rap1b | 0.112 | ndufa4l2a | -0.046 |
| si:dkey-238i5.2 | 0.111 | rpl39 | -0.045 |
| si:ch211-156j16.1 | 0.110 | esama | -0.045 |
| vat1 | 0.110 | nasp | -0.044 |
| flot2a | 0.110 | dut | -0.044 |
| lama5 | 0.109 | kcnj8 | -0.044 |
| cst14a.2 | 0.108 | gngt2b | -0.044 |
| flot1b | 0.108 | rps29 | -0.044 |
| cd81a | 0.107 | epas1a | -0.044 |
| tmem88b | 0.106 | emx2 | -0.043 |
| tpm4a | 0.106 | zgc:195173 | -0.043 |