solute carrier family 12 member 8
ZFIN 
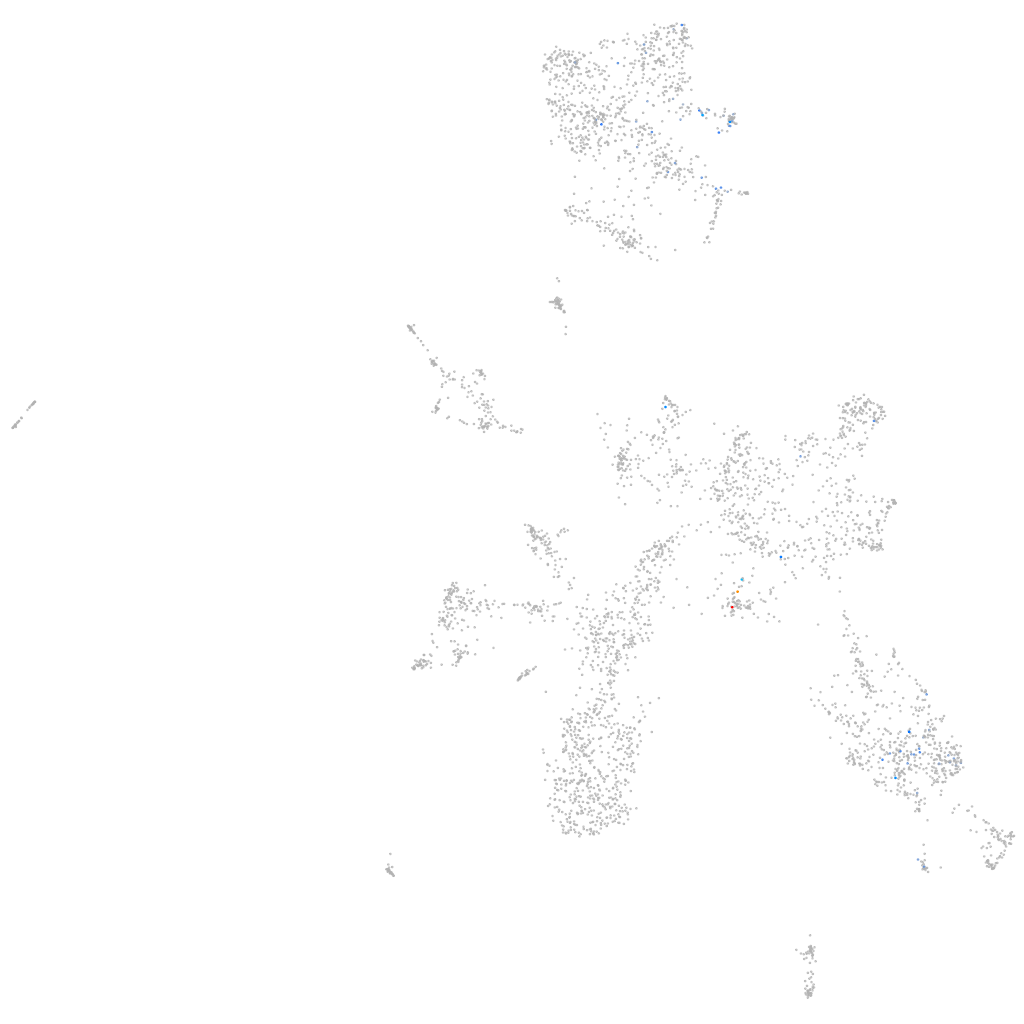
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC101882729 | 0.436 | h3f3d | -0.089 |
| pimr186 | 0.412 | COX3 | -0.084 |
| si:ch73-281f12.4 | 0.399 | ubc | -0.071 |
| si:dkey-1h24.2 | 0.397 | ptmaa | -0.068 |
| si:dkey-190j3.4 | 0.308 | si:ch73-1a9.3 | -0.058 |
| kita | 0.276 | dynll1 | -0.056 |
| irx3a | 0.266 | hmgb3a | -0.054 |
| si:ch73-266f23.1 | 0.251 | hmgn6 | -0.053 |
| LOC101883513 | 0.233 | hmgn7 | -0.052 |
| si:dkey-65b12.6 | 0.228 | marcksl1b | -0.051 |
| thap3 | 0.224 | NC-002333.17 | -0.051 |
| XLOC-041601 | 0.221 | ywhabl | -0.050 |
| atg4b | 0.214 | calm3b | -0.049 |
| setx | 0.213 | h2afx1 | -0.049 |
| si:dkey-57h18.1 | 0.210 | oaz1b | -0.048 |
| gfpt2 | 0.206 | ptmab | -0.047 |
| creb3l1 | 0.200 | si:ch1073-429i10.3.1 | -0.046 |
| mlh3 | 0.199 | hnrnpa0a | -0.044 |
| peli2 | 0.197 | eif1b | -0.044 |
| s100s | 0.197 | mt-atp6 | -0.044 |
| agr2 | 0.193 | tuba1c | -0.042 |
| snrpd3 | 0.190 | insm1a | -0.042 |
| usp16 | 0.189 | jpt1b | -0.041 |
| gxylt2 | 0.187 | rtn1a | -0.041 |
| ern2 | 0.185 | elavl3 | -0.041 |
| si:ch211-163l21.8 | 0.179 | f11r.1 | -0.041 |
| cd164 | 0.172 | khdrbs1a | -0.041 |
| mtfr1 | 0.169 | tmeff1b | -0.041 |
| foxa3 | 0.164 | csnk2b | -0.040 |
| PAQR9 | 0.163 | gng5 | -0.040 |
| zgc:162964 | 0.161 | drap1 | -0.040 |
| si:ch211-265o23.1 | 0.159 | cox7b | -0.039 |
| rrbp1b | 0.159 | calm2a | -0.039 |
| ppcs | 0.156 | paip2b | -0.039 |
| nectin4a | 0.154 | rnasekb | -0.038 |