solute carrier family 12 member 2
ZFIN 
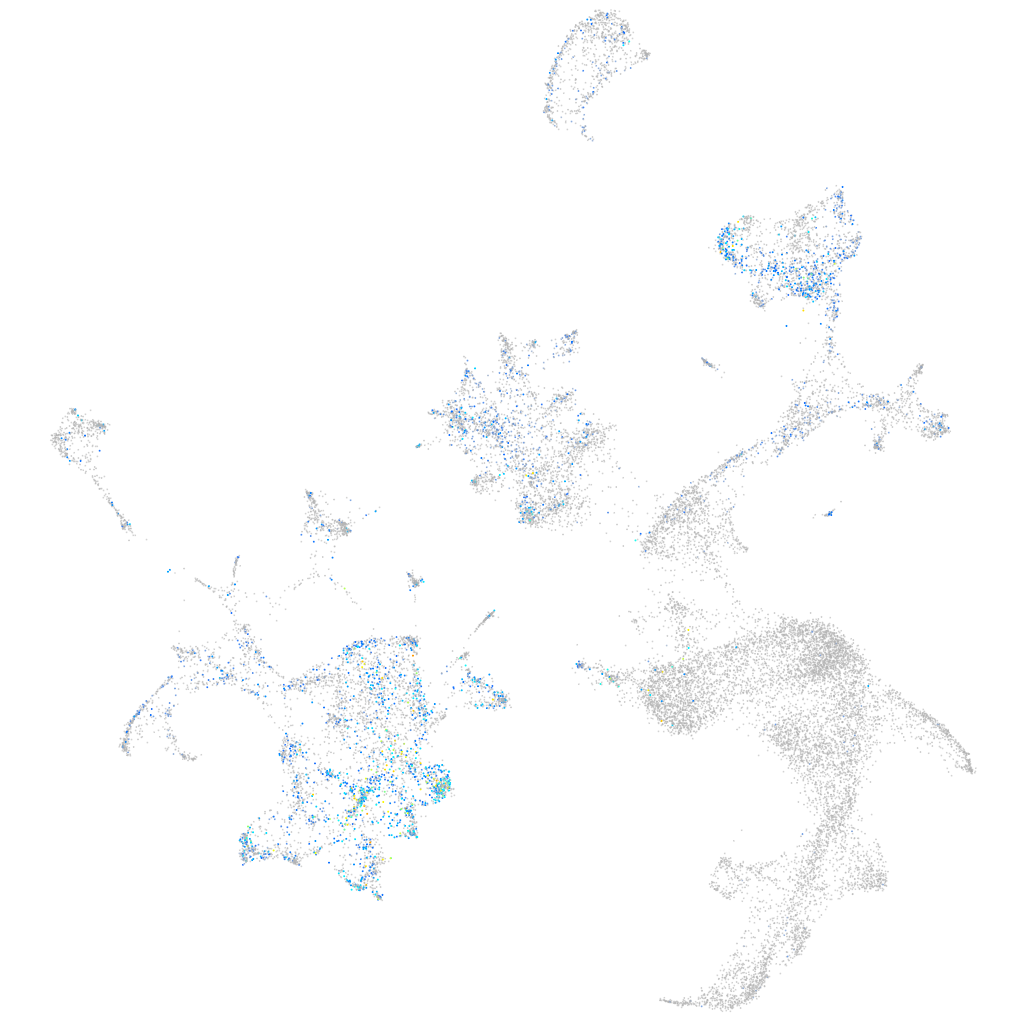
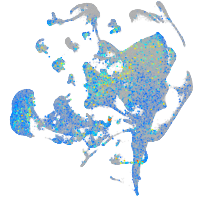
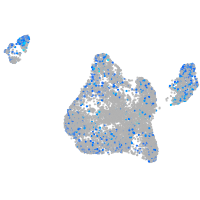
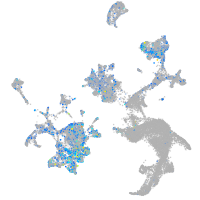
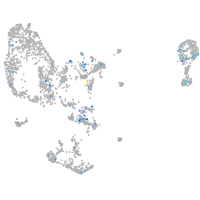
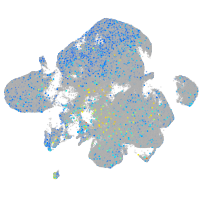
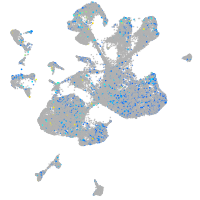
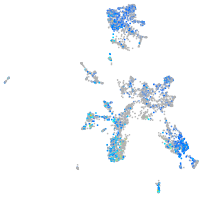
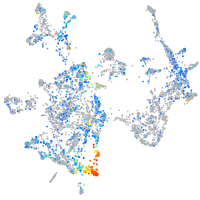
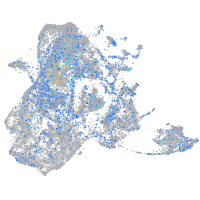
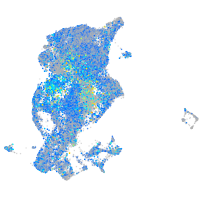
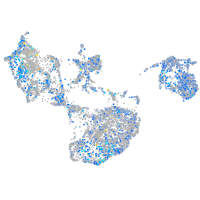
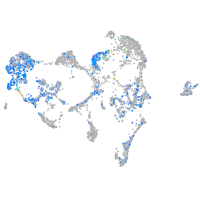
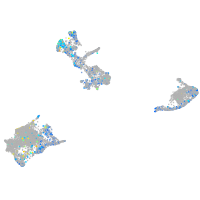
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scarb2a | 0.303 | hbae1.1 | -0.235 |
| col4a1 | 0.300 | hbae1.3 | -0.231 |
| podxl | 0.296 | hbae3 | -0.224 |
| vat1 | 0.295 | hbbe1.3 | -0.221 |
| plvapb | 0.289 | hbbe2 | -0.221 |
| plecb | 0.288 | cahz | -0.218 |
| cdh5 | 0.287 | hbbe1.1 | -0.216 |
| kdrl | 0.278 | hemgn | -0.207 |
| igfbp7 | 0.278 | prdx2 | -0.204 |
| mcamb | 0.275 | hbbe1.2 | -0.200 |
| krt8 | 0.263 | fth1a | -0.199 |
| si:ch211-156j16.1 | 0.262 | blvrb | -0.196 |
| cd81a | 0.259 | alas2 | -0.196 |
| plpp3 | 0.258 | slc4a1a | -0.187 |
| si:dkey-261h17.1 | 0.256 | nt5c2l1 | -0.180 |
| si:dkeyp-97a10.2 | 0.251 | tspo | -0.180 |
| slc9a3r1b | 0.251 | zgc:163057 | -0.179 |
| ahnak | 0.250 | si:ch211-250g4.3 | -0.172 |
| krt18a.1 | 0.249 | epb41b | -0.171 |
| cpn1 | 0.248 | creg1 | -0.170 |
| marcksl1a | 0.247 | nmt1b | -0.161 |
| tcima | 0.246 | si:ch211-207c6.2 | -0.158 |
| akap12b | 0.245 | plac8l1 | -0.152 |
| msna | 0.245 | tmod4 | -0.150 |
| gnai2a | 0.244 | hbbe3 | -0.139 |
| kdr | 0.243 | hbae5 | -0.136 |
| cldn5b | 0.243 | si:ch211-227m13.1 | -0.134 |
| dock6 | 0.240 | sptb | -0.133 |
| si:ch211-145b13.6 | 0.239 | tfr1a | -0.133 |
| tmsb4x | 0.239 | uros | -0.131 |
| arhgap31 | 0.239 | zgc:56095 | -0.130 |
| she | 0.237 | tmem14ca | -0.127 |
| cav1 | 0.236 | rfesd | -0.126 |
| tmem108 | 0.235 | hdr | -0.125 |
| tspan4b | 0.234 | klf1 | -0.124 |