SIX homeobox 4b
ZFIN 
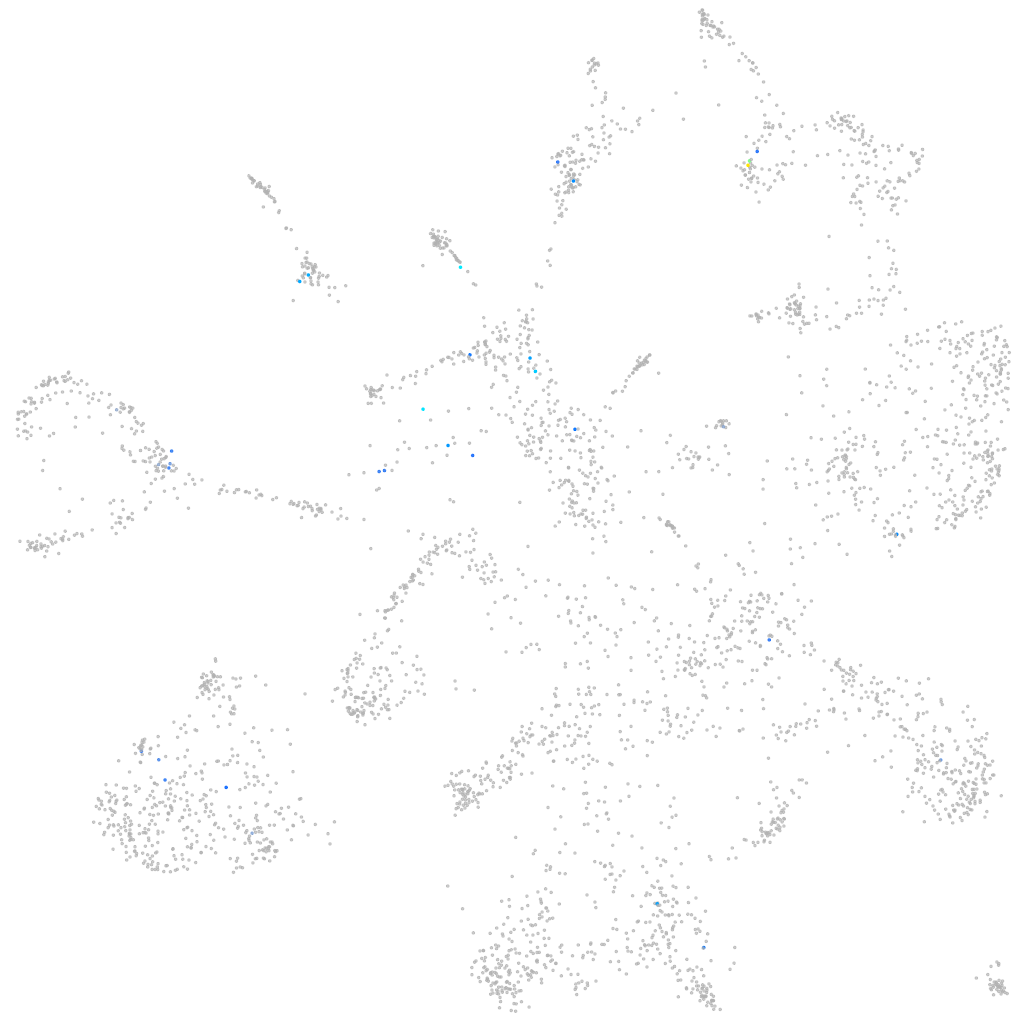
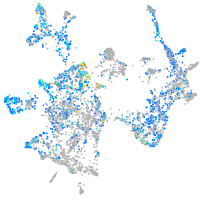
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC108192140 | 0.363 | rpl18 | -0.050 |
| CR392341.1 | 0.346 | rpl30 | -0.047 |
| cx28.6 | 0.343 | rps13 | -0.040 |
| BX072576.1 | 0.277 | aoc2 | -0.040 |
| CU929391.1 | 0.272 | nkx2.3 | -0.040 |
| CABZ01043166.1 | 0.270 | rps26l | -0.038 |
| FP102171.1 | 0.249 | rpl5b | -0.037 |
| slc15a1a | 0.243 | rpl23 | -0.036 |
| CABZ01074708.1 | 0.239 | XLOC-041870 | -0.036 |
| nrn1la | 0.231 | rpl12 | -0.035 |
| XLOC-001661 | 0.229 | ctsz | -0.035 |
| CABZ01056826.1 | 0.219 | cavin2b | -0.035 |
| BX323801.1 | 0.213 | rps15a | -0.034 |
| CR788226.1 | 0.211 | stau2 | -0.034 |
| epha8 | 0.209 | mdka | -0.033 |
| bscl2l | 0.203 | rps8a | -0.033 |
| si:ch211-108c6.2 | 0.203 | col6a2 | -0.032 |
| CR769777.2 | 0.198 | bambia | -0.032 |
| otop2 | 0.188 | ier2a | -0.030 |
| ftr22 | 0.188 | podxl | -0.030 |
| CU074330.1 | 0.175 | cebpb | -0.030 |
| agmo | 0.173 | map2 | -0.030 |
| amfrb | 0.172 | tcea1 | -0.029 |
| rxfp3.3a3 | 0.169 | scaf1 | -0.029 |
| XLOC-044291 | 0.166 | rps2 | -0.029 |
| fgf18a | 0.163 | ppfibp1a | -0.029 |
| XLOC-007078 | 0.162 | tmem98 | -0.029 |
| mfsd6l | 0.161 | COLEC10 | -0.029 |
| slc17a5 | 0.157 | col6a1 | -0.027 |
| akap12a | 0.155 | foxf1 | -0.027 |
| LOC100537272 | 0.153 | rpl18a | -0.027 |
| ryr1a | 0.151 | sertad2a | -0.027 |
| cdh17 | 0.151 | si:dkey-195m11.8 | -0.027 |
| zgc:153759 | 0.150 | rps3 | -0.027 |
| capn15 | 0.150 | bmp2a | -0.027 |