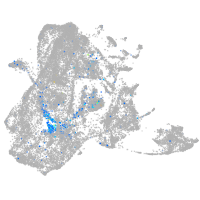
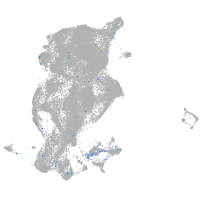
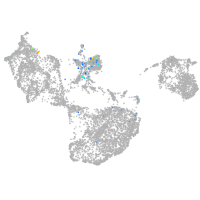
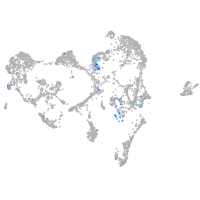
SIX homeobox 3a
ZFIN 
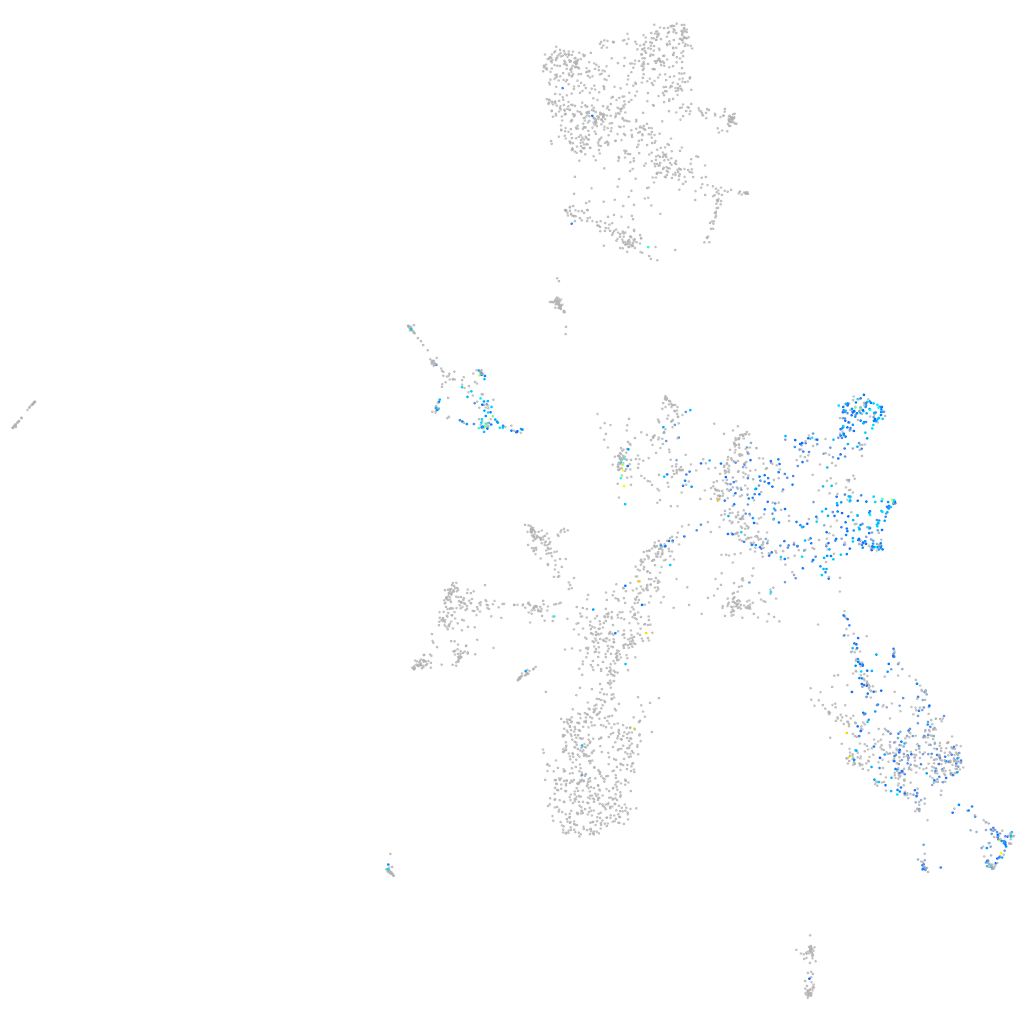
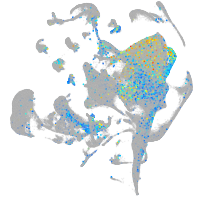
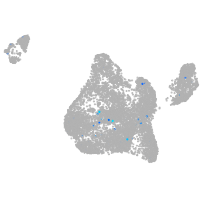
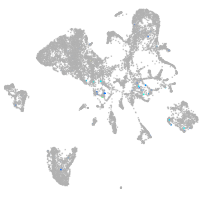
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| six3b | 0.491 | cldnh | -0.307 |
| id1 | 0.393 | anxa5b | -0.291 |
| nid2a | 0.377 | txn | -0.277 |
| dlx5a | 0.374 | gstp1 | -0.259 |
| si:dkey-261h17.1 | 0.355 | bik | -0.255 |
| fgf24 | 0.351 | calm1a | -0.248 |
| six1b | 0.349 | vamp2 | -0.241 |
| si:ch211-243g18.2 | 0.343 | calb2a | -0.239 |
| dlx3b | 0.338 | tuba1c | -0.230 |
| nanos1 | 0.338 | calm1b | -0.230 |
| fgf8a | 0.334 | rnasekb | -0.228 |
| sp8a | 0.326 | eml2 | -0.224 |
| tpm4a | 0.326 | ponzr6 | -0.218 |
| col14a1a | 0.318 | rtn1a | -0.209 |
| her6 | 0.315 | gipc2 | -0.206 |
| dag1 | 0.314 | bdnf | -0.199 |
| six4b | 0.310 | tspan2a | -0.197 |
| akap12b | 0.309 | gng13a | -0.194 |
| sp8b | 0.304 | ip6k2a | -0.192 |
| her12 | 0.303 | scg2a | -0.191 |
| lamb1a | 0.300 | kcnc1a | -0.191 |
| XLOC-016198 | 0.295 | prr15la | -0.191 |
| sox3 | 0.293 | atp6v1g1 | -0.190 |
| krt18b | 0.282 | gabarapa | -0.190 |
| cx43.4 | 0.278 | tprg1 | -0.190 |
| dmrt3a | 0.278 | atp6v1e1b | -0.189 |
| otx1 | 0.277 | selenow2b | -0.189 |
| cnn2 | 0.277 | LOC100536671 | -0.186 |
| cnn3a | 0.276 | gnb1a | -0.183 |
| col1a2 | 0.275 | ptmaa | -0.183 |
| rac1a | 0.275 | gbgt1l4 | -0.182 |
| her15.1 | 0.274 | atp6v0cb | -0.181 |
| pak2a | 0.273 | tnrc6c1 | -0.180 |
| sept2 | 0.273 | icn | -0.180 |
| qkia | 0.270 | tmem59 | -0.180 |