SIN3-HDAC complex associated factor
ZFIN 
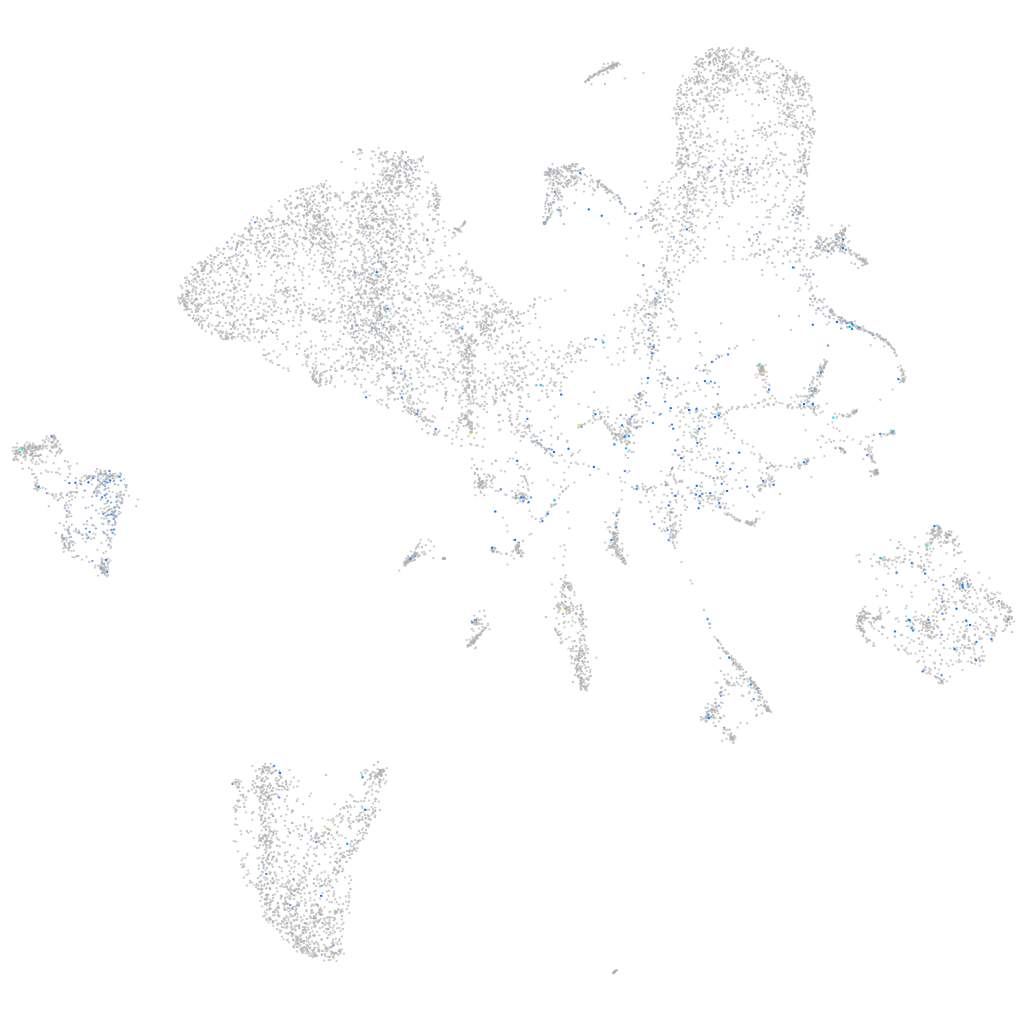
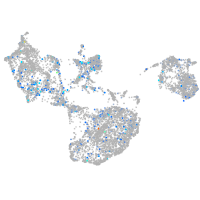
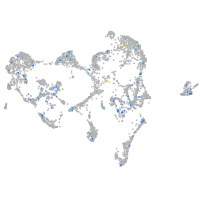
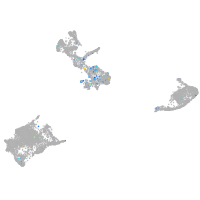
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC101882800 | 0.162 | gamt | -0.126 |
| BX908803.2 | 0.154 | gapdh | -0.117 |
| mst1rb | 0.143 | gatm | -0.115 |
| XLOC-039631 | 0.138 | bhmt | -0.104 |
| sult1st7 | 0.134 | apoa4b.1 | -0.103 |
| XLOC-026746 | 0.133 | fbp1b | -0.100 |
| LOC103911313 | 0.133 | apoc2 | -0.099 |
| epcam | 0.126 | scp2a | -0.099 |
| si:ch211-222l21.1 | 0.125 | afp4 | -0.099 |
| her9 | 0.123 | apoa1b | -0.097 |
| si:dkey-61f9.2 | 0.123 | suclg1 | -0.097 |
| cldnb | 0.122 | agxtb | -0.095 |
| CU929199.1 | 0.121 | glud1b | -0.095 |
| ubc | 0.119 | gpx4a | -0.093 |
| LOC101882128 | 0.119 | pnp4b | -0.091 |
| hmgn2 | 0.118 | pgm1 | -0.091 |
| cldn7b | 0.117 | mat1a | -0.090 |
| mrvi1 | 0.115 | grhprb | -0.087 |
| h3f3d | 0.115 | apoa2 | -0.087 |
| inhbb | 0.114 | cx32.3 | -0.087 |
| khdrbs1a | 0.113 | apobb.1 | -0.085 |
| btg2 | 0.112 | apom | -0.085 |
| si:ch211-223a21.3 | 0.112 | fabp10a | -0.085 |
| si:dkey-83f18.8 | 0.112 | serpina1 | -0.085 |
| si:dkey-28d5.9 | 0.112 | gnmt | -0.085 |
| crocc2 | 0.111 | rbp2b | -0.084 |
| si:ch211-63p21.4 | 0.111 | aldob | -0.084 |
| hmgb1a | 0.110 | ahcy | -0.084 |
| si:ch73-1a9.3 | 0.110 | hao1 | -0.084 |
| ptmab | 0.109 | suclg2 | -0.084 |
| ly6m2 | 0.108 | tfa | -0.084 |
| LOC101884879 | 0.108 | dap | -0.083 |
| atxn7l1 | 0.107 | serpina1l | -0.083 |
| ywhaz | 0.107 | zgc:123103 | -0.083 |
| cirbpb | 0.107 | ambp | -0.082 |