si:rp71-1k13.6
ZFIN 
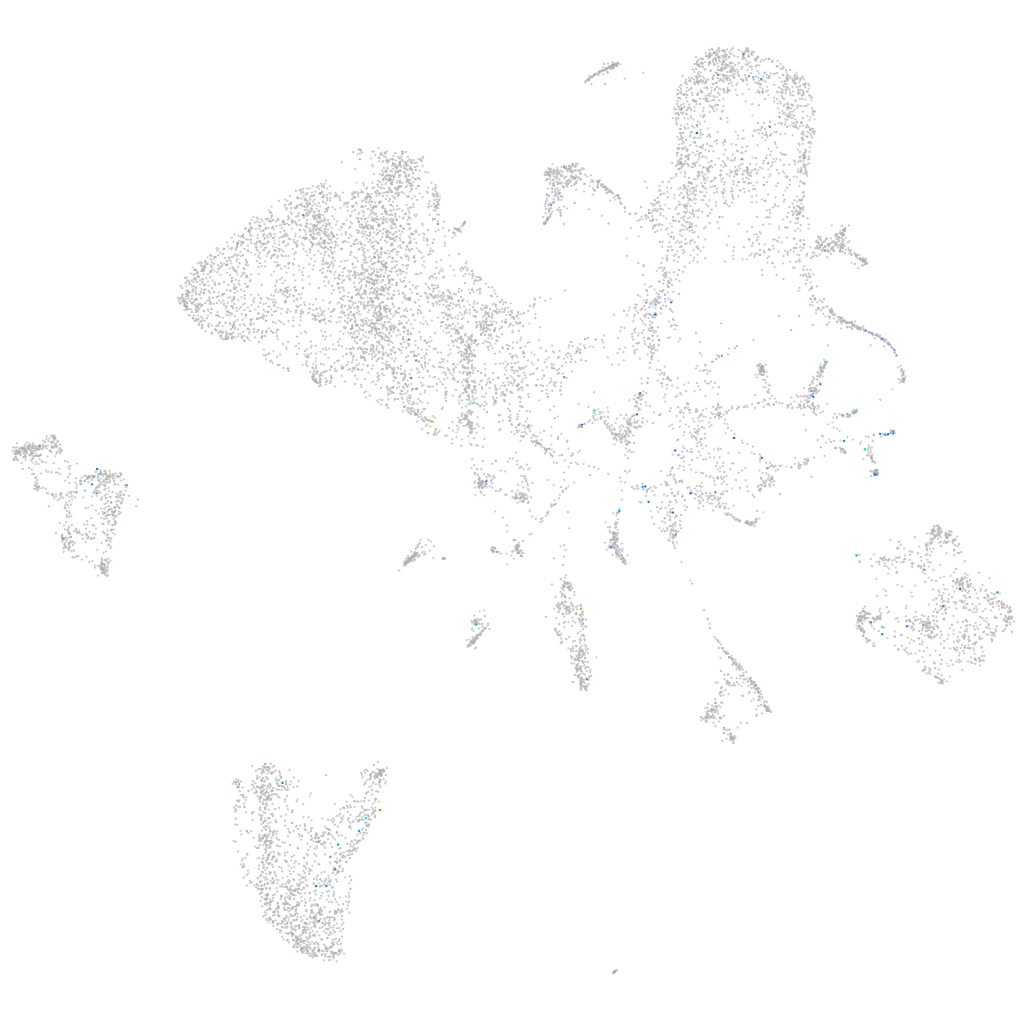
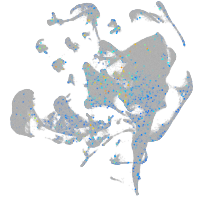
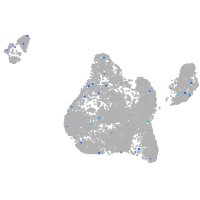
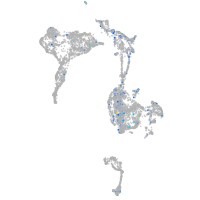
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC110438300 | 0.265 | gamt | -0.048 |
| XLOC-036220 | 0.245 | apoa1b | -0.044 |
| igf1 | 0.232 | agxtb | -0.044 |
| rad21l1 | 0.220 | apoc1 | -0.043 |
| cnga3b | 0.207 | apoa2 | -0.042 |
| AL929276.2 | 0.205 | fetub | -0.042 |
| LOC110437996 | 0.201 | serpina1l | -0.042 |
| LOC101886621 | 0.196 | apom | -0.041 |
| ptpdc1b | 0.189 | mat1a | -0.041 |
| LOC103912007 | 0.167 | fabp10a | -0.040 |
| CR352243.1 | 0.167 | apoc2 | -0.040 |
| LOC101885528.1 | 0.161 | rbp4 | -0.040 |
| zgc:123321 | 0.156 | pnp4b | -0.040 |
| BX571828.1 | 0.153 | ces2 | -0.040 |
| plcxd1 | 0.153 | serpina1 | -0.040 |
| LO017815.1 | 0.150 | ambp | -0.040 |
| FP325130.1 | 0.150 | bhmt | -0.040 |
| itpr1b | 0.145 | LOC110437731 | -0.039 |
| zgc:113223 | 0.142 | si:dkey-86l18.10 | -0.039 |
| LOC101883201 | 0.139 | phyhd1 | -0.039 |
| LO017829.1 | 0.138 | kng1 | -0.039 |
| fitm1 | 0.135 | tfa | -0.039 |
| BX908744.2 | 0.135 | fgg | -0.039 |
| pnpla2 | 0.131 | apoa4b.1 | -0.039 |
| CABZ01070631.1 | 0.119 | agxta | -0.038 |
| CABZ01052573.1 | 0.118 | hpda | -0.038 |
| nobox | 0.114 | zgc:123103 | -0.038 |
| BX248497.1 | 0.111 | rbp2b | -0.037 |
| mhc1uka | 0.111 | ftcd | -0.037 |
| LO018363.2 | 0.109 | ttc36 | -0.037 |
| CT573139.2 | 0.108 | afp4 | -0.037 |
| prkcea | 0.105 | nipsnap3a | -0.037 |
| svep1 | 0.105 | grhprb | -0.037 |
| si:ch73-206p6.1 | 0.103 | c9 | -0.037 |
| mtnr1ab | 0.102 | comtd1 | -0.036 |