si:dkeyp-87d8.8
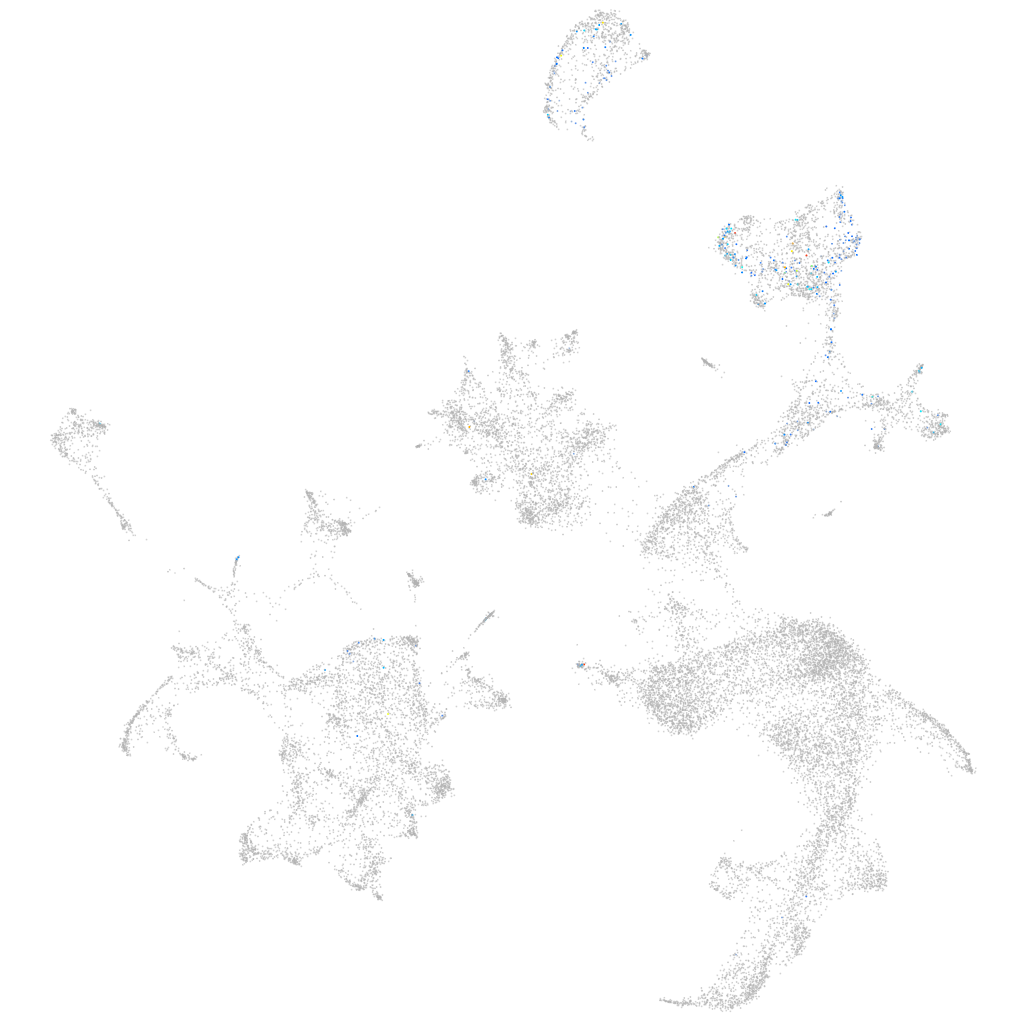
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mpeg1.1 | 0.187 | hbae1.1 | -0.088 |
| si:ch211-212k18.7 | 0.179 | hbbe1.3 | -0.088 |
| havcr1 | 0.176 | hbbe2 | -0.087 |
| slc7a7 | 0.173 | hbae3 | -0.086 |
| CU499330.1 | 0.172 | hbae1.3 | -0.084 |
| cmklr1 | 0.171 | cahz | -0.081 |
| fcer1gl | 0.168 | hbbe1.1 | -0.081 |
| lgals3bpb | 0.167 | hemgn | -0.075 |
| ptpn6 | 0.165 | hbbe1.2 | -0.074 |
| mfap4 | 0.164 | zgc:56095 | -0.072 |
| cx32.2 | 0.163 | fth1a | -0.072 |
| laptm5 | 0.163 | alas2 | -0.070 |
| LOC108180725 | 0.162 | si:ch211-250g4.3 | -0.069 |
| lgals2a | 0.162 | blvrb | -0.065 |
| LOC101886104 | 0.161 | nt5c2l1 | -0.064 |
| ctsc | 0.161 | slc4a1a | -0.063 |
| si:ch211-106a19.1 | 0.160 | lmo2 | -0.063 |
| ptprc | 0.157 | creg1 | -0.062 |
| ctss2.2 | 0.157 | zgc:163057 | -0.061 |
| sult3st4 | 0.157 | si:ch211-207c6.2 | -0.060 |
| cxcr3.2 | 0.157 | epb41b | -0.057 |
| si:dkey-5n18.1 | 0.156 | tmod4 | -0.056 |
| spi1b | 0.155 | tspo | -0.055 |
| coro1a | 0.155 | aqp1a.1 | -0.055 |
| irf8 | 0.155 | nmt1b | -0.051 |
| cebpb | 0.154 | uros | -0.051 |
| cyba | 0.154 | plac8l1 | -0.050 |
| gm2a | 0.154 | hbae5 | -0.050 |
| tspan36 | 0.154 | sptb | -0.049 |
| tnfb | 0.151 | si:ch211-227m13.1 | -0.048 |
| si:zfos-741a10.3 | 0.149 | tfr1a | -0.047 |
| tcirg1b | 0.149 | rfesd | -0.047 |
| arpc1b | 0.149 | dmtn | -0.046 |
| si:zfos-1069f5.1 | 0.148 | add2 | -0.046 |
| si:ch73-158p21.3 | 0.148 | histh1l | -0.045 |