si:dkeyp-82a1.2
ZFIN 
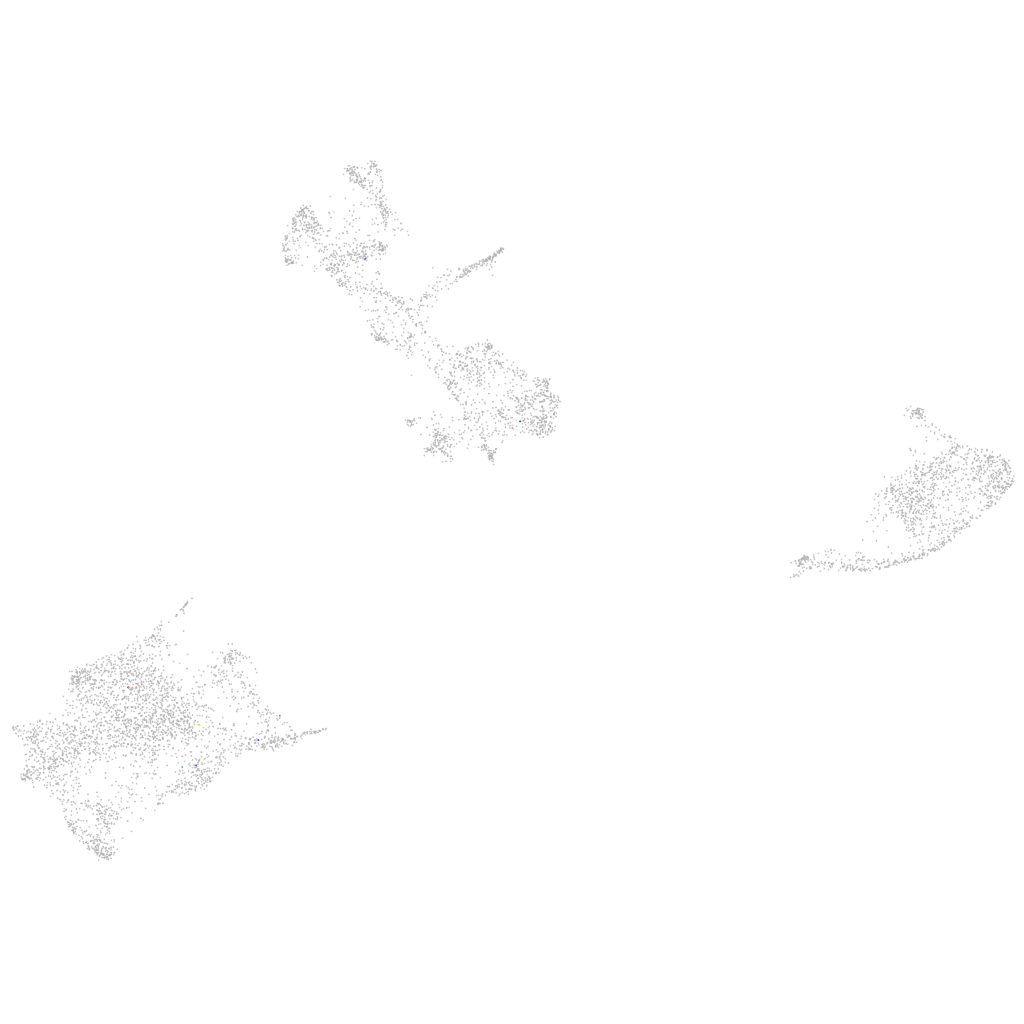
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 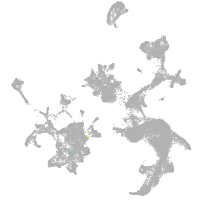 |
pronephros 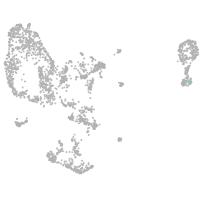 |
neural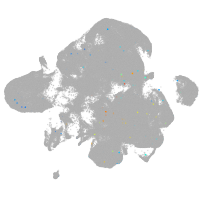 |
spinal cord / glia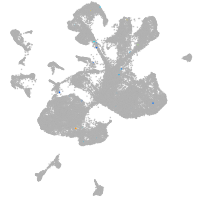 |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| XLOC-018977 | 0.597 | rpl10 | -0.043 |
| LOC108179274 | 0.394 | rps5 | -0.035 |
| btr01 | 0.392 | rpl14 | -0.034 |
| cplx3b | 0.378 | rps4x | -0.033 |
| XLOC-032159 | 0.359 | fabp3 | -0.030 |
| lim2.1 | 0.355 | rpl7 | -0.028 |
| XLOC-029303 | 0.280 | rpl30 | -0.027 |
| gfra3 | 0.269 | rplp0 | -0.027 |
| slitrk3a | 0.266 | rpl22 | -0.026 |
| nkx1.2lb | 0.264 | rps16 | -0.025 |
| si:dkey-242h9.3 | 0.251 | naca | -0.025 |
| pcdh8 | 0.251 | rps28 | -0.025 |
| parm1 | 0.231 | rplp2 | -0.024 |
| dthd1 | 0.230 | uba52 | -0.023 |
| RASSF5 | 0.219 | rps15a | -0.023 |
| chodl | 0.210 | ran | -0.022 |
| calca | 0.196 | rps17 | -0.020 |
| cryba1l1 | 0.191 | eef1b2 | -0.020 |
| si:ch211-161c3.5 | 0.185 | mt-co1 | -0.020 |
| ssh2a | 0.176 | ctsla | -0.019 |
| LOC108179143 | 0.175 | NC-002333.17 | -0.019 |
| csrp3 | 0.166 | ubb | -0.019 |
| LOC100333572 | 0.161 | vamp3 | -0.019 |
| dusp26 | 0.158 | zgc:153867 | -0.019 |
| c1qtnf4 | 0.152 | atp5pf | -0.019 |
| tmem102 | 0.142 | s100a10b | -0.019 |
| BX510337.1 | 0.140 | fthl27 | -0.018 |
| nyap2a | 0.139 | rps26 | -0.018 |
| tnfb | 0.135 | aldh9a1a.1 | -0.018 |
| ggcta | 0.129 | rps3 | -0.018 |
| kdelc1 | 0.128 | rpl5b | -0.018 |
| irx3a | 0.127 | eif3f | -0.017 |
| ift20 | 0.127 | tpt1 | -0.017 |
| prep | 0.125 | pnrc2 | -0.016 |
| tppp3 | 0.123 | cox6a1 | -0.016 |