si:dkeyp-69c1.9
ZFIN 
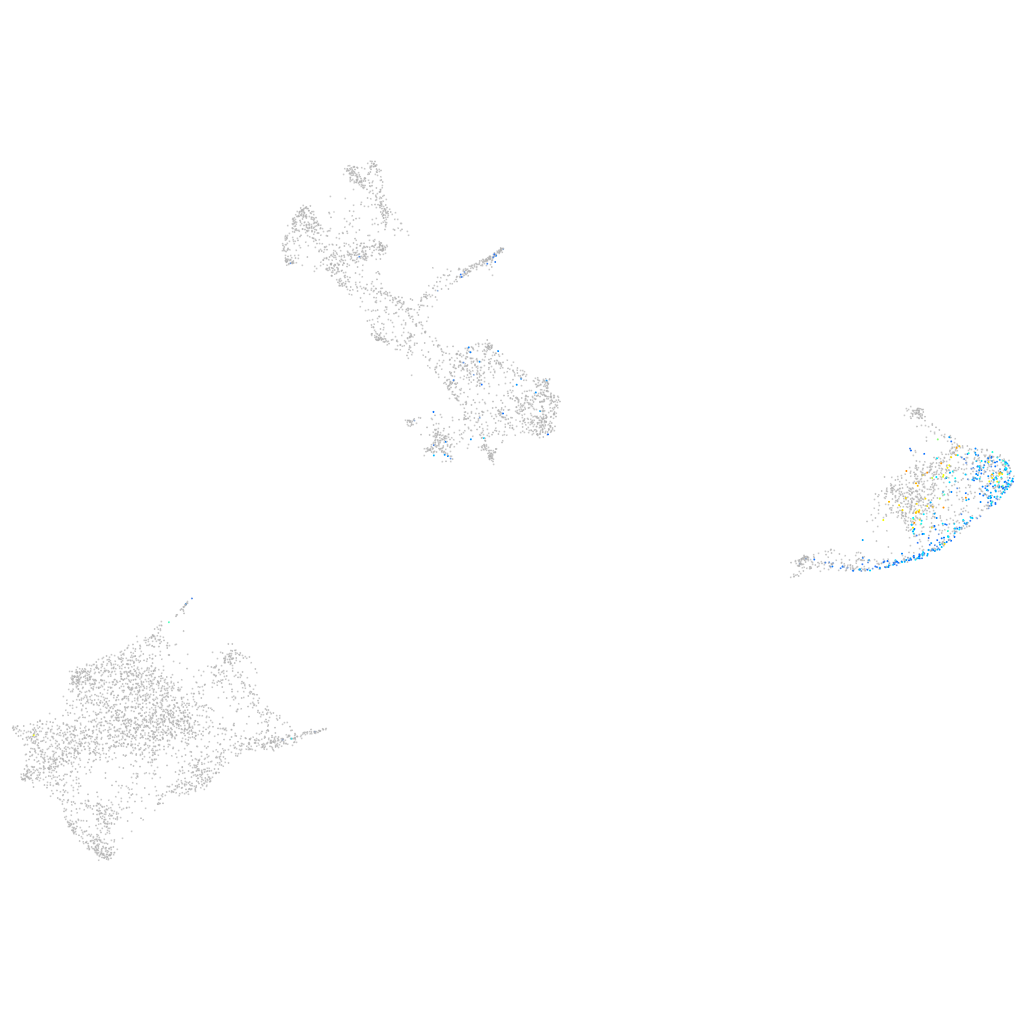
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tyrp1b | 0.419 | paics | -0.225 |
| tyrp1a | 0.414 | CABZ01021592.1 | -0.220 |
| dct | 0.414 | uraha | -0.182 |
| pmela | 0.408 | si:dkey-251i10.2 | -0.175 |
| zgc:91968 | 0.391 | tmem130 | -0.163 |
| si:ch73-389b16.1 | 0.388 | slc2a15a | -0.154 |
| oca2 | 0.385 | mdh1aa | -0.153 |
| slc24a5 | 0.381 | atic | -0.150 |
| kita | 0.355 | phyhd1 | -0.150 |
| mtbl | 0.349 | impdh1b | -0.146 |
| prkar1b | 0.348 | glulb | -0.145 |
| slc22a2 | 0.345 | aox5 | -0.143 |
| tyr | 0.336 | si:ch211-251b21.1 | -0.139 |
| aadac | 0.332 | prps1a | -0.131 |
| slc39a10 | 0.331 | bscl2l | -0.124 |
| slc45a2 | 0.324 | pax7b | -0.120 |
| gstt1a | 0.306 | cyb5a | -0.118 |
| kcnj13 | 0.306 | pax7a | -0.114 |
| anxa1a | 0.300 | bco1 | -0.113 |
| timeless | 0.300 | sprb | -0.112 |
| hsd20b2 | 0.298 | ppat | -0.111 |
| lamp1a | 0.298 | gmps | -0.108 |
| SPAG9 | 0.290 | sult1st1 | -0.108 |
| spra | 0.289 | aldob | -0.107 |
| slc29a3 | 0.282 | krt18b | -0.104 |
| slc37a2 | 0.280 | cx30.3 | -0.104 |
| zdhhc2 | 0.279 | prdx5 | -0.103 |
| atp6v0a2b | 0.279 | cax1 | -0.103 |
| slc3a2a | 0.278 | mibp | -0.101 |
| si:ch211-195b13.1 | 0.277 | si:ch211-194k22.8 | -0.100 |
| gpr61 | 0.276 | prdx1 | -0.100 |
| tspan10 | 0.272 | TMEM19 | -0.099 |
| mchr2 | 0.272 | aqp3a | -0.098 |
| mlpha | 0.271 | shmt1 | -0.098 |
| tfap2e | 0.269 | oacyl | -0.097 |