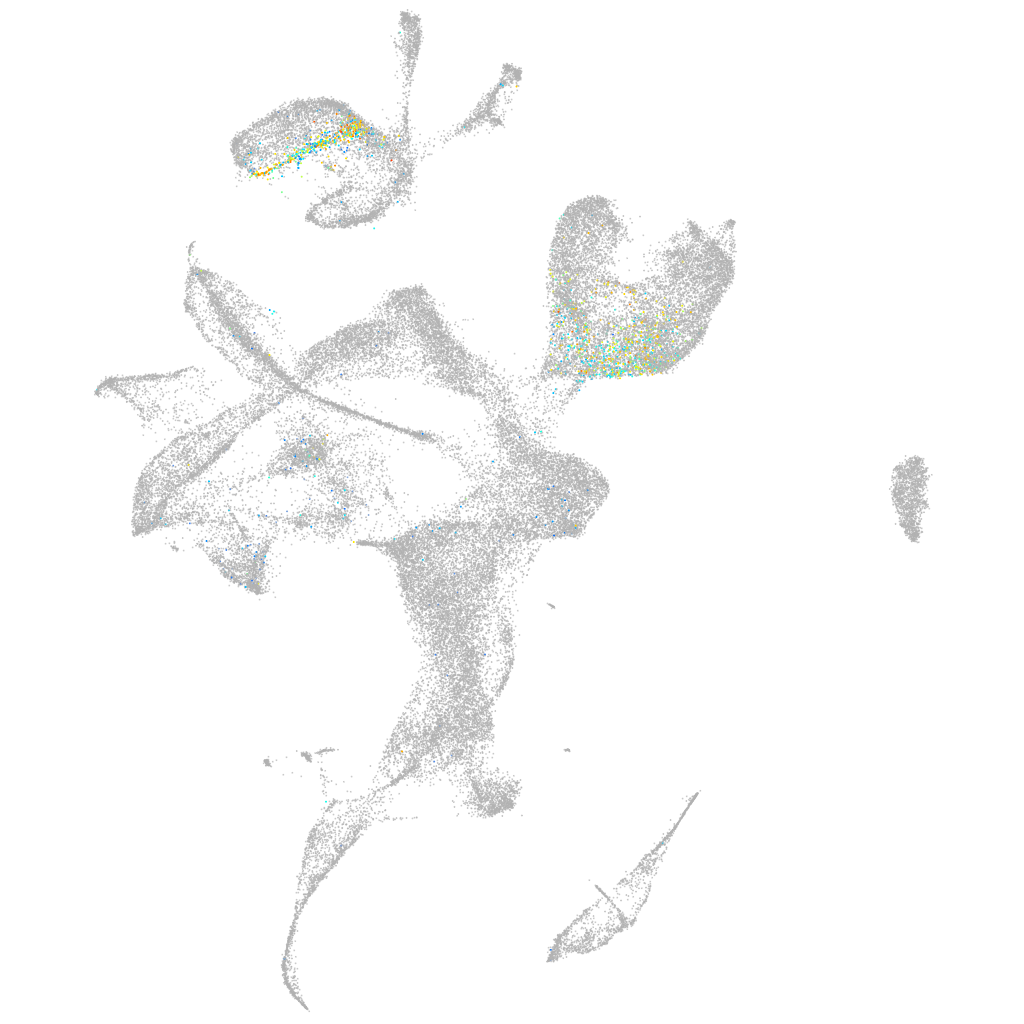si:dkeyp-41f9.4
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| chgb | 0.292 | msi1 | -0.092 |
| bhlhe22 | 0.226 | hmgb2a | -0.090 |
| bhlhe23 | 0.210 | pcna | -0.089 |
| meis2b | 0.207 | msna | -0.088 |
| prdm8b | 0.200 | si:ch73-281n10.2 | -0.088 |
| nacad | 0.192 | stmn1a | -0.085 |
| CU928046.1 | 0.163 | tuba8l4 | -0.083 |
| dpysl4 | 0.162 | dut | -0.083 |
| LOC101884359 | 0.162 | lbr | -0.081 |
| FP102783.1 | 0.161 | selenoh | -0.080 |
| otofa | 0.160 | rrm1 | -0.079 |
| foxp2 | 0.157 | mki67 | -0.079 |
| pax10 | 0.148 | ccnd1 | -0.078 |
| lhx2b | 0.145 | si:ch211-137a8.4 | -0.078 |
| ret | 0.140 | banf1 | -0.076 |
| rorb | 0.140 | nutf2l | -0.075 |
| psd2 | 0.138 | chaf1a | -0.075 |
| gpm6ab | 0.138 | ccna2 | -0.073 |
| scrt2 | 0.137 | dek | -0.073 |
| nol4la | 0.137 | tuba8l | -0.073 |
| pcbp3 | 0.135 | rpa3 | -0.072 |
| crip3 | 0.135 | fen1 | -0.071 |
| CR356233.1 | 0.135 | nasp | -0.071 |
| gpm6aa | 0.134 | mcm7 | -0.070 |
| gnb3a | 0.128 | cks1b | -0.069 |
| si:ch211-204a13.2 | 0.127 | zbtb18 | -0.068 |
| zgc:194629 | 0.126 | lig1 | -0.068 |
| pbx1a | 0.122 | rrm2 | -0.068 |
| glrba | 0.120 | her15.1 | -0.068 |
| NAV1 (1 of many) | 0.118 | mibp | -0.067 |
| rims2a | 0.118 | CABZ01005379.1 | -0.067 |
| gbx2 | 0.118 | cldn5a | -0.066 |
| rai14 | 0.116 | id1 | -0.066 |
| bmpr2a | 0.115 | rpa2 | -0.065 |
| dscama | 0.115 | si:ch211-156b7.4 | -0.064 |