si:dkey-81l17.6
ZFIN 
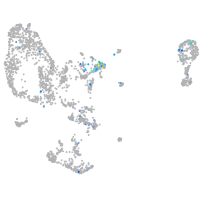
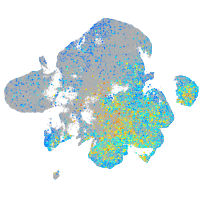
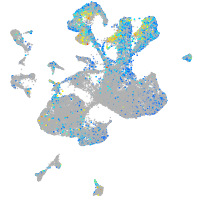
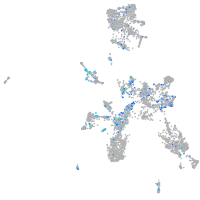
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| stmn1b | 0.229 | aldob | -0.093 |
| nova2 | 0.190 | ahcy | -0.090 |
| myt1la | 0.187 | fbp1b | -0.088 |
| XLOC-019901 | 0.187 | gapdh | -0.087 |
| gng3 | 0.182 | aldh6a1 | -0.086 |
| elavl3 | 0.176 | eef1da | -0.085 |
| csdc2a | 0.176 | mat1a | -0.084 |
| rtn1b | 0.175 | eno3 | -0.084 |
| dhrs7ca | 0.175 | gamt | -0.084 |
| grapb | 0.173 | nupr1b | -0.084 |
| arf3b | 0.167 | cx32.3 | -0.083 |
| CABZ01046949.1 | 0.166 | scp2a | -0.079 |
| gfra4a | 0.162 | glud1b | -0.075 |
| tuba1c | 0.162 | suclg1 | -0.075 |
| pimr94 | 0.161 | sod1 | -0.075 |
| cnrip1a | 0.158 | pgm1 | -0.075 |
| zc4h2 | 0.156 | gatm | -0.074 |
| SAMD12 | 0.155 | agxtb | -0.074 |
| apc2 | 0.155 | gstt1a | -0.073 |
| sncb | 0.154 | abat | -0.071 |
| si:dkey-276j7.1 | 0.153 | gnmt | -0.071 |
| tmeff1b | 0.152 | gpx4a | -0.070 |
| tuba1a | 0.150 | cat | -0.070 |
| gpm6aa | 0.150 | aldh7a1 | -0.070 |
| c1qa | 0.149 | pnp4b | -0.070 |
| tubb5 | 0.149 | ckba | -0.070 |
| pcdh2g12 | 0.149 | pklr | -0.070 |
| trim46b | 0.149 | dap | -0.070 |
| celf4 | 0.148 | tdo2a | -0.069 |
| elavl4 | 0.146 | agxta | -0.069 |
| mir129-1 | 0.146 | fabp10a | -0.068 |
| hmgb3a | 0.143 | slco1d1 | -0.068 |
| nos1apb | 0.143 | gstz1 | -0.067 |
| cspg5a | 0.142 | hpda | -0.067 |
| foxp2 | 0.142 | slc25a34 | -0.067 |