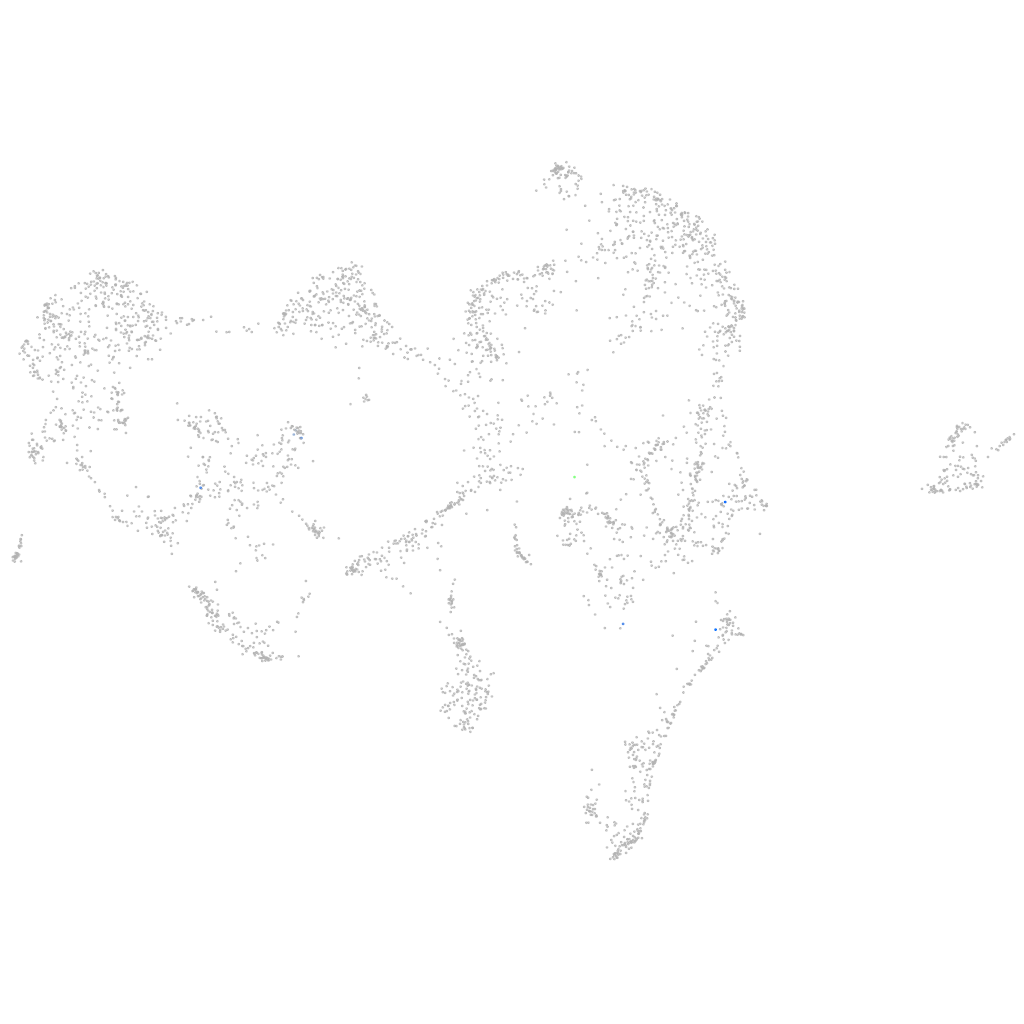
si:dkey-7f16.3
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle 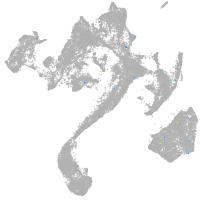 |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural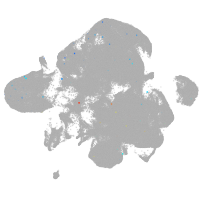 |
spinal cord / glia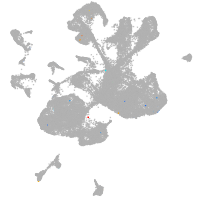 |
eye |
taste / olfactory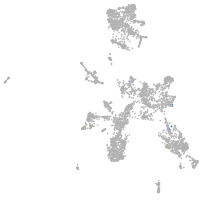 |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| AL954359.1 | 0.797 | cldnh | -0.040 |
| LOC100331404 | 0.615 | krt91 | -0.035 |
| grm2b | 0.559 | cox5ab | -0.034 |
| LOC100331412 | 0.520 | eef1da | -0.034 |
| wscd1b | 0.465 | chmp4bb | -0.028 |
| spef2 | 0.450 | rwdd1 | -0.027 |
| aatka | 0.449 | ier2b | -0.027 |
| bgnb | 0.424 | ezrb | -0.027 |
| si:dkey-98f17.5 | 0.420 | tmem258 | -0.026 |
| XLOC-008233 | 0.413 | zgc:110333 | -0.026 |
| ttyh1 | 0.389 | prdx3 | -0.026 |
| FAT4 | 0.364 | capns1a | -0.026 |
| ca4a | 0.360 | malb | -0.026 |
| si:dkey-110g7.8 | 0.353 | rpl6 | -0.026 |
| slitrk2 | 0.335 | ahcy | -0.026 |
| dydc2 | 0.333 | mdh2 | -0.025 |
| FO744854.1 | 0.314 | actc1b | -0.025 |
| CR385035.1 | 0.312 | icn2 | -0.025 |
| gsx1 | 0.301 | tdh | -0.025 |
| ppic | 0.301 | nipsnap2 | -0.025 |
| gpr37l1b | 0.293 | krt5 | -0.024 |
| cyp26b1 | 0.292 | rnf128a | -0.024 |
| pimr80 | 0.289 | dnaja2a | -0.024 |
| slc1a2b | 0.276 | cox6c | -0.024 |
| pimr209 | 0.275 | gbgt1l4 | -0.024 |
| rhot1b | 0.265 | gars | -0.024 |
| ak8 | 0.264 | fbp1a | -0.024 |
| auts2b | 0.263 | atp6v1h | -0.024 |
| birc6-as2 | 0.259 | hbbe1.3 | -0.024 |
| znf644b | 0.259 | mtch2 | -0.024 |
| tbc1d31 | 0.259 | si:ch211-202f5.3 | -0.023 |
| kbtbd11 | 0.257 | rab5c | -0.023 |
| samd1b | 0.256 | si:ch211-166a6.5 | -0.023 |
| atp1a1b | 0.246 | dap | -0.023 |
| unk | 0.244 | zgc:56525 | -0.023 |