si:dkey-79d12.5
ZFIN 
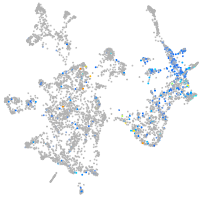
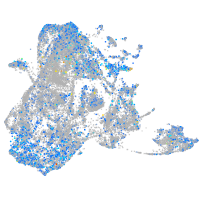
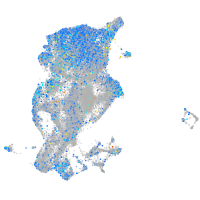
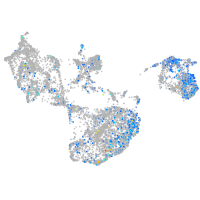
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC110440040 | 0.174 | hbbe1.3 | -0.055 |
| cuzd1.2 | 0.168 | hbae3 | -0.050 |
| ccr10 | 0.157 | hbae1.1 | -0.046 |
| LOC100535070 | 0.147 | gstp1 | -0.044 |
| AL935044.2 | 0.147 | tnni2a.4 | -0.040 |
| FO904966.1 | 0.147 | tmsb4x | -0.040 |
| CABZ01046996.1 | 0.144 | hbbe1.1 | -0.040 |
| arhgef38 | 0.144 | rbp4l | -0.038 |
| socs3a | 0.138 | hbbe1.2 | -0.037 |
| FAM107A (1 of many) | 0.138 | actc1b | -0.034 |
| CABZ01072523.1 | 0.138 | ckma | -0.034 |
| si:dkey-88l16.2 | 0.138 | tyrp1a | -0.033 |
| vps9d1 | 0.135 | cyt1l | -0.033 |
| si:dkey-121n8.7 | 0.133 | tuba1c | -0.031 |
| LOC103909475 | 0.132 | mllt11 | -0.031 |
| f3b | 0.130 | dct | -0.031 |
| si:ch211-207b24.4 | 0.128 | vat1 | -0.030 |
| si:dkeyp-100a1.6 | 0.128 | fabp7a | -0.030 |
| btg2 | 0.127 | cotl1 | -0.030 |
| AL928908.2 | 0.125 | pvalb1 | -0.030 |
| pim1 | 0.124 | tyrp1b | -0.029 |
| noctb | 0.124 | oca2 | -0.028 |
| gadd45ga | 0.123 | gstt1a | -0.027 |
| LOC101883052.1 | 0.121 | gpd1b | -0.027 |
| BX248082.1 | 0.120 | pvalb2 | -0.027 |
| AL732567.1 | 0.120 | krt4 | -0.027 |
| sdc4 | 0.120 | ttn.1 | -0.026 |
| marco | 0.117 | gch2 | -0.026 |
| CU929070.1 | 0.117 | prdx1 | -0.026 |
| trpm1b | 0.116 | atp2a1 | -0.026 |
| CR383676.1 | 0.113 | hbbe2 | -0.026 |
| pprc1 | 0.113 | elavl3 | -0.026 |
| CR388148.2 | 0.113 | prkag2a | -0.026 |
| ier2b | 0.111 | gap43 | -0.026 |
| ddit3 | 0.108 | stxbp1a | -0.026 |