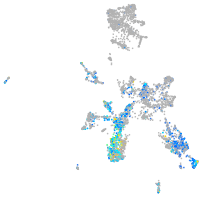
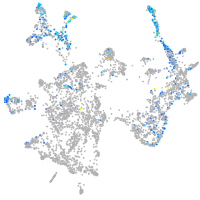
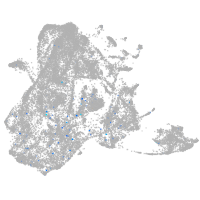
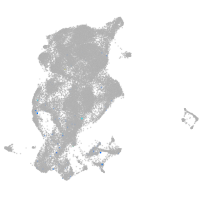
si:dkey-70b23.2
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| daw1 | 0.162 | ttn.1 | -0.028 |
| CR450782.1 | 0.150 | actc1b | -0.027 |
| hid1b | 0.145 | pvalb2 | -0.026 |
| ccdc173 | 0.122 | ttn.2 | -0.026 |
| nitr12 | 0.117 | mybphb | -0.026 |
| LOC100329490 | 0.109 | pvalb1 | -0.025 |
| si:dkeyp-110a12.4 | 0.106 | si:ch73-367p23.2 | -0.025 |
| mapk15 | 0.105 | mylz3 | -0.025 |
| ttll2 | 0.105 | mylpfb | -0.025 |
| hepacama | 0.102 | tnni2a.4 | -0.024 |
| slc6a11b | 0.100 | pabpc4 | -0.024 |
| ftr12 | 0.100 | ak1 | -0.024 |
| XLOC-006319 | 0.099 | tnnt3b | -0.024 |
| XLOC-003690 | 0.096 | mylpfa | -0.024 |
| smkr1 | 0.094 | ckmb | -0.024 |
| capslb | 0.093 | acta1b | -0.024 |
| XLOC-023308 | 0.091 | tnnc2 | -0.024 |
| nme5 | 0.090 | tpma | -0.024 |
| bicc2 | 0.088 | tnnt3a | -0.024 |
| ccdc151 | 0.087 | atp2a1 | -0.024 |
| CABZ01012962.1 | 0.085 | gamt | -0.023 |
| siglec15l | 0.085 | gatm | -0.023 |
| ect2l | 0.085 | cfl2 | -0.023 |
| CABZ01052527.1 | 0.084 | si:ch211-255p10.3 | -0.023 |
| ccdc114 | 0.083 | gapdh | -0.023 |
| CU074421.1 | 0.081 | myoz1b | -0.023 |
| ankrd45 | 0.081 | aldoab | -0.023 |
| zgc:165461 | 0.081 | eef1da | -0.023 |
| si:ch211-77g15.32 | 0.080 | myl1 | -0.023 |
| si:dkey-148f10.4 | 0.079 | myom2a | -0.023 |
| AL935199.1 | 0.079 | casq1b | -0.023 |
| si:ch211-248e11.2 | 0.078 | rbfox1l | -0.023 |
| ak8 | 0.077 | tmem38a | -0.023 |
| si:ch211-66e2.5 | 0.077 | atp1a2a | -0.022 |
| her4.4 | 0.077 | fxr2 | -0.022 |