si:dkey-69o16.5
ZFIN 
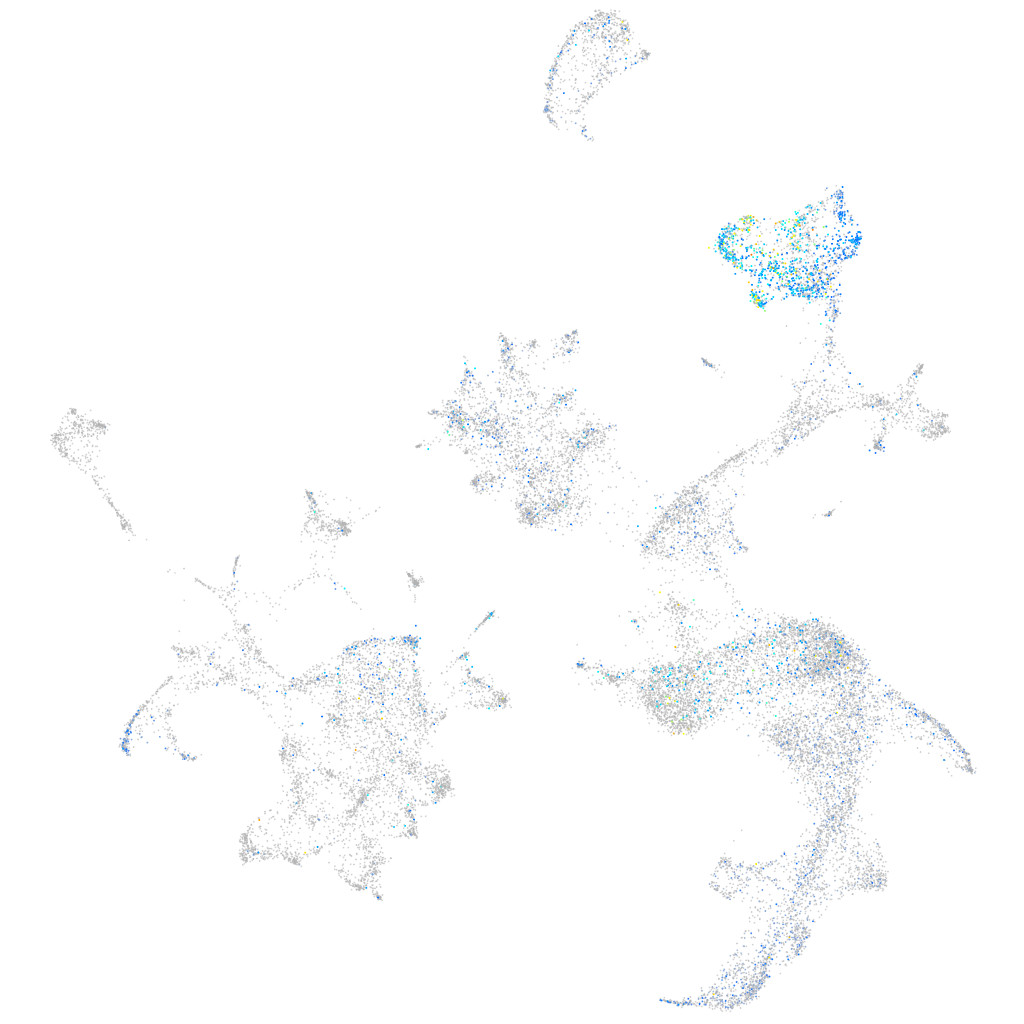
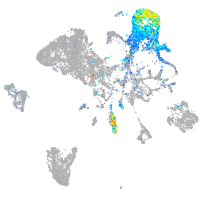
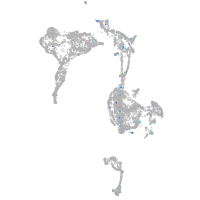
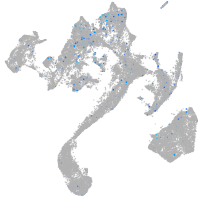
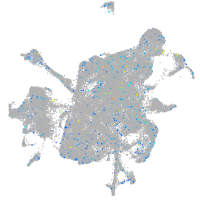
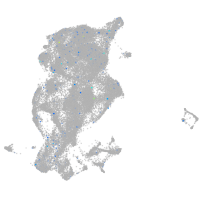
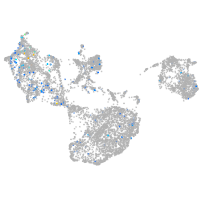
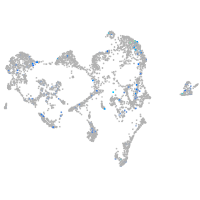
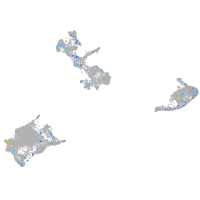
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| havcr1 | 0.453 | si:ch211-222l21.1 | -0.110 |
| si:dkey-5n18.1 | 0.444 | si:dkey-261h17.1 | -0.100 |
| lgals2a | 0.439 | akap12b | -0.099 |
| cmklr1 | 0.418 | krt18a.1 | -0.096 |
| mpeg1.1 | 0.415 | jpt1b | -0.094 |
| mfap4 | 0.404 | lmo2 | -0.091 |
| slc3a2a | 0.402 | aqp1a.1 | -0.090 |
| ccl34b.1 | 0.401 | hbae1.1 | -0.090 |
| ctss2.2 | 0.388 | cdh5 | -0.090 |
| lgals3bpb | 0.386 | sox7 | -0.089 |
| tspan36 | 0.385 | tpm4a | -0.088 |
| slc7a7 | 0.380 | hmgb1b | -0.088 |
| cx32.2 | 0.377 | fth1a | -0.086 |
| ctsl.1 | 0.372 | clec14a | -0.086 |
| tnfsf12 | 0.369 | slc4a1a | -0.086 |
| ctsba | 0.369 | si:ch211-156j16.1 | -0.086 |
| rnaset2l | 0.365 | hbae1.3 | -0.085 |
| si:zfos-1069f5.1 | 0.363 | hbbe1.3 | -0.084 |
| marco | 0.362 | cldn5b | -0.082 |
| irf8 | 0.359 | etv2 | -0.082 |
| mpp1 | 0.353 | she | -0.081 |
| ctsc | 0.351 | qkia | -0.081 |
| lygl1 | 0.351 | hbbe2 | -0.081 |
| tnfb | 0.349 | egfl7 | -0.080 |
| gpd1a | 0.345 | hbbe1.1 | -0.080 |
| si:ch211-147m6.1 | 0.344 | hbae3 | -0.080 |
| cxcr3.2 | 0.343 | cnn3a | -0.079 |
| c1qc | 0.338 | marcksb | -0.079 |
| CU499330.1 | 0.337 | col4a1 | -0.079 |
| gm2a | 0.337 | crip2 | -0.078 |
| lgals9l1 | 0.335 | igfbp7 | -0.078 |
| si:ch211-212k18.7 | 0.335 | fscn1a | -0.078 |
| cndp2 | 0.334 | hbbe1.2 | -0.078 |
| si:ch211-194m7.3 | 0.334 | nova2 | -0.077 |
| ms4a17a.10 | 0.332 | tmem88a | -0.077 |