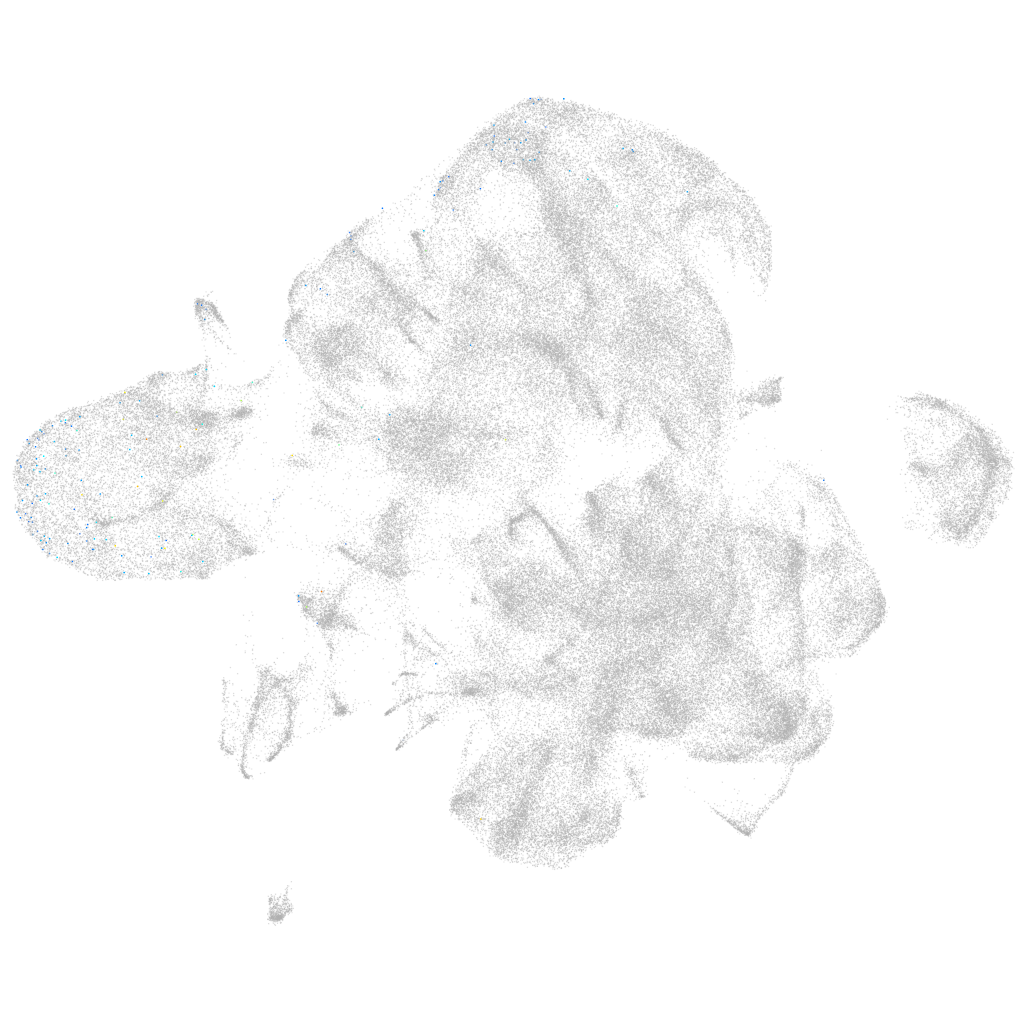
si:dkey-46g23.2
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs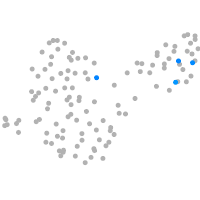 |
endoderm |
axial mesoderm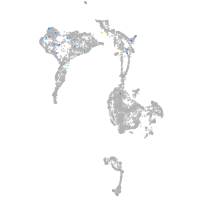 |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 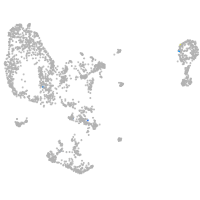 |
neural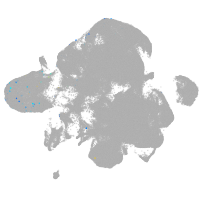 |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line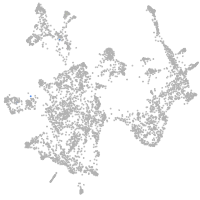 |
epidermis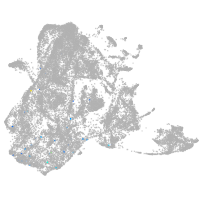 |
periderm |
fin |
ionocytes / mucous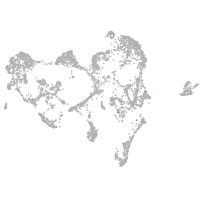 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| zgc:171446 | 0.144 | ptmaa | -0.043 |
| si:dkey-46g23.5 | 0.087 | rpl37 | -0.042 |
| smtla | 0.084 | rps10 | -0.040 |
| zgc:158445 | 0.077 | zgc:114188 | -0.039 |
| ovgp1 | 0.076 | h3f3a | -0.038 |
| pou5f3 | 0.074 | tuba1c | -0.036 |
| polr3gla | 0.069 | CR383676.1 | -0.035 |
| add3b | 0.065 | marcksl1a | -0.033 |
| hspb1 | 0.065 | elavl3 | -0.033 |
| stm | 0.065 | ppiab | -0.031 |
| si:ch211-152c2.3 | 0.063 | hnrnpa0l | -0.031 |
| zgc:56699 | 0.062 | fabp3 | -0.031 |
| s100a1 | 0.058 | gpm6aa | -0.031 |
| FERMT3 | 0.056 | rtn1a | -0.030 |
| apoeb | 0.056 | tmsb4x | -0.029 |
| lrwd1 | 0.056 | stmn1b | -0.028 |
| si:ch1073-80i24.3 | 0.055 | nova2 | -0.028 |
| cyp2k22 | 0.055 | rps17 | -0.027 |
| apela | 0.055 | si:ch1073-429i10.3.1 | -0.027 |
| dkc1 | 0.054 | tmsb | -0.026 |
| aldob | 0.054 | fam168a | -0.026 |
| vrtn | 0.054 | mt-atp6 | -0.025 |
| rrp15 | 0.053 | zgc:158463 | -0.025 |
| vox | 0.053 | actc1b | -0.025 |
| si:dkey-228b2.6 | 0.053 | celf2 | -0.025 |
| foxd5 | 0.053 | COX3 | -0.024 |
| crabp2b | 0.052 | gpm6ab | -0.024 |
| asb11 | 0.052 | pvalb1 | -0.024 |
| foxh1 | 0.052 | rnasekb | -0.024 |
| nnr | 0.052 | ppdpfb | -0.023 |
| fbl | 0.052 | h3f3c | -0.023 |
| pprc1 | 0.052 | mt-nd1 | -0.023 |
| ccdc137 | 0.051 | nme2b.1 | -0.023 |
| ved | 0.051 | ckbb | -0.022 |
| bms1 | 0.051 | calm1a | -0.022 |