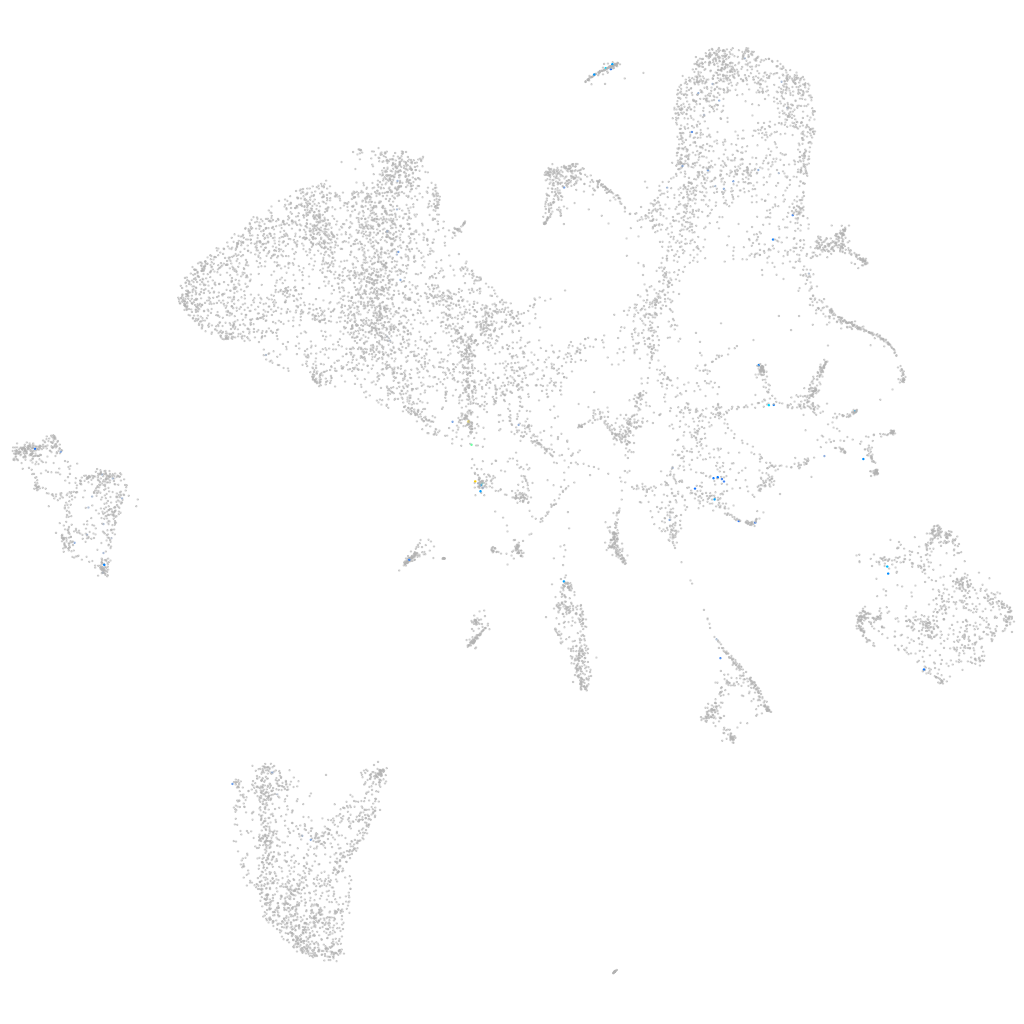
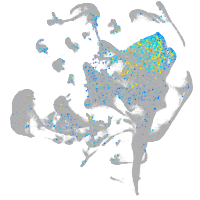
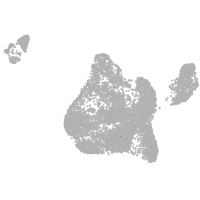
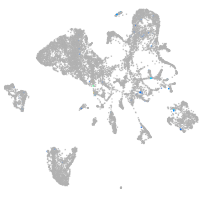
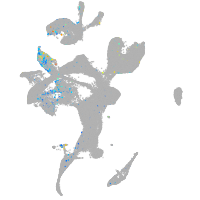
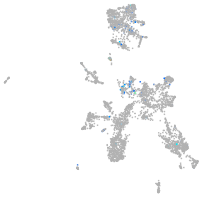
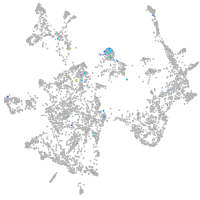
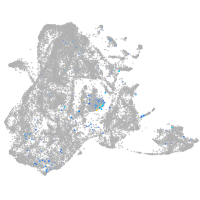
si:dkey-35i13.1
ZFIN 
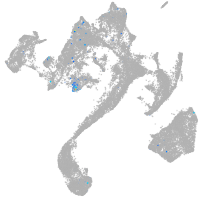
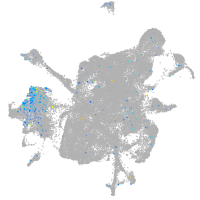
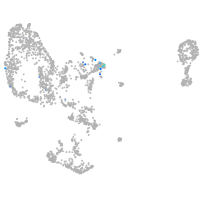
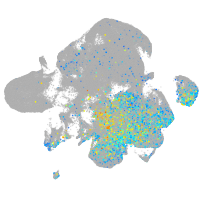
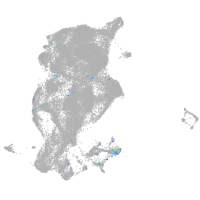
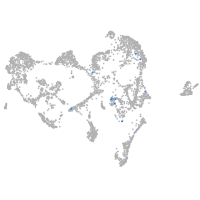
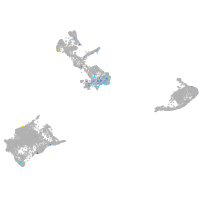
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gpr176 | 0.345 | aldh6a1 | -0.050 |
| LOC100332354 | 0.303 | gapdh | -0.046 |
| LOC101883911 | 0.300 | ahcy | -0.046 |
| cilp2 | 0.281 | rps18 | -0.046 |
| AL954831.1 | 0.265 | gamt | -0.045 |
| msnb | 0.265 | gatm | -0.041 |
| si:ch211-257p13.3 | 0.235 | ssr2 | -0.041 |
| trpm1a | 0.229 | pnp4b | -0.040 |
| dchs1a | 0.225 | aldh7a1 | -0.040 |
| tmem136b | 0.222 | gstr | -0.039 |
| sycp1 | 0.222 | scp2a | -0.038 |
| inpp5kb | 0.214 | mat1a | -0.038 |
| tacr3a | 0.214 | atp5mc3b | -0.038 |
| CT573799.2 | 0.212 | chchd10 | -0.037 |
| zgc:153760 | 0.198 | agxta | -0.037 |
| fam155a | 0.197 | cx32.3 | -0.037 |
| mitfa | 0.196 | cebpd | -0.037 |
| nrg3b | 0.191 | fbp1b | -0.037 |
| pcolceb | 0.190 | mgst1.2 | -0.037 |
| trhde.2 | 0.189 | eno3 | -0.037 |
| gpr186 | 0.186 | tfa | -0.037 |
| olfcd3 | 0.186 | nipsnap3a | -0.036 |
| CU856222.1 | 0.185 | isoc2 | -0.036 |
| erfl1 | 0.184 | sdr16c5b | -0.036 |
| CT573433.1 | 0.182 | ckba | -0.036 |
| LOC108179567 | 0.180 | upb1 | -0.036 |
| sulf2b | 0.179 | gstt1a | -0.035 |
| pbx3b | 0.179 | etnppl | -0.035 |
| LOC101884756 | 0.174 | tdo2a | -0.035 |
| gabra2a | 0.174 | ttr | -0.035 |
| ppfia2 | 0.171 | hpda | -0.035 |
| si:ch73-127m5.1 | 0.170 | fabp10a | -0.035 |
| myo16 | 0.168 | ttc36 | -0.035 |
| CU681836.1 | 0.168 | uox | -0.035 |
| chst8 | 0.168 | hspd1 | -0.035 |