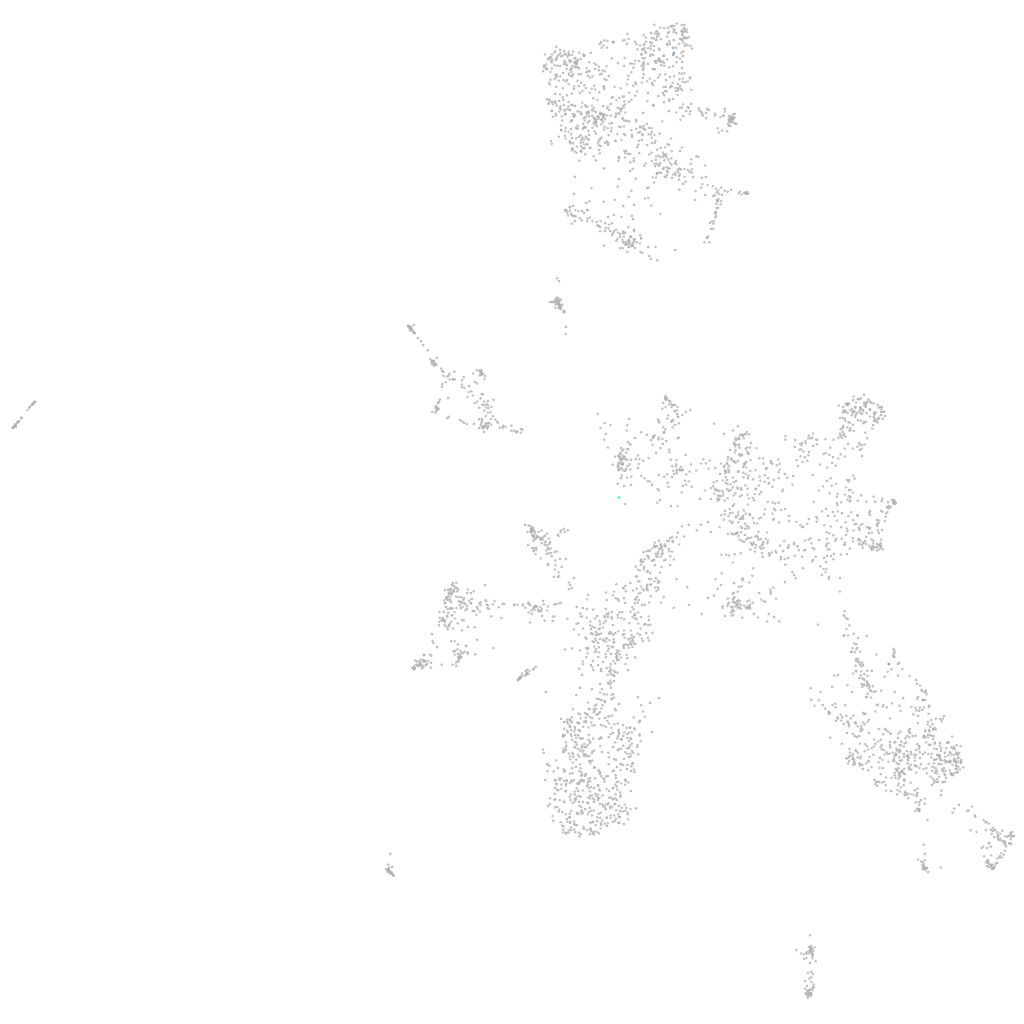
si:dkey-30c15.13
ZFIN 
Other cell groups


all cells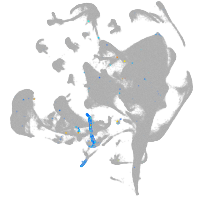 |
blastomeres |
PGCs |
endoderm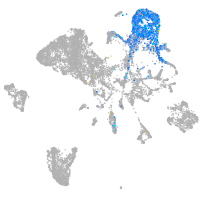 |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural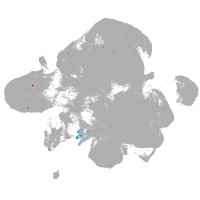 |
spinal cord / glia |
eye |
taste / olfactory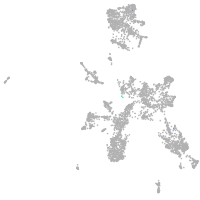 |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous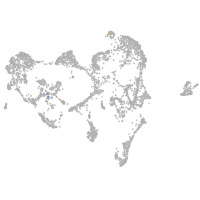 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC103911551 | 0.908 | fkbp1aa | -0.036 |
| si:ch211-152c8.5 | 0.740 | si:ch1073-429i10.3.1 | -0.035 |
| miox | 0.719 | rpl23a | -0.034 |
| cmtm8b | 0.661 | pabpc1a | -0.033 |
| LOC103912005 | 0.601 | slc25a3b | -0.032 |
| BX539307.1 | 0.601 | NC-002333.17 | -0.030 |
| eomesa | 0.598 | rps17 | -0.030 |
| si:dkeyp-41f9.4 | 0.516 | epcam | -0.030 |
| ccl33.3 | 0.506 | serf2 | -0.027 |
| XLOC-040753 | 0.445 | hmgb2a | -0.027 |
| apela | 0.366 | atp5fa1 | -0.027 |
| gucy2d | 0.364 | serbp1a | -0.027 |
| neurod6a | 0.353 | zgc:56493 | -0.025 |
| slc8a1a | 0.343 | calm1a | -0.025 |
| ncs1b | 0.315 | ndufb3 | -0.025 |
| b3gat2 | 0.298 | atp1b1a | -0.024 |
| BX957317.2 | 0.298 | edf1 | -0.024 |
| tbr1a | 0.297 | gapdhs | -0.024 |
| gabra5 | 0.296 | gnb1b | -0.024 |
| LOC100535941 | 0.286 | rps18 | -0.023 |
| CDHR1 (1 of many) | 0.269 | rpl5b | -0.023 |
| crygmx | 0.261 | vdac3 | -0.023 |
| birc6-as2 | 0.257 | atp6v1e1b | -0.023 |
| GPR139 | 0.254 | skp1 | -0.023 |
| CR786577.1 | 0.241 | psmb2 | -0.022 |
| ehbp1l1b | 0.224 | itm2ba | -0.022 |
| apba2a | 0.222 | atp5mf | -0.022 |
| dlgap3 | 0.221 | bcl2l10 | -0.022 |
| upb1 | 0.217 | cox5aa | -0.022 |
| slc25a37 | 0.214 | sem1 | -0.022 |
| zgc:85722 | 0.198 | gnb1a | -0.021 |
| lepr | 0.196 | ssr2 | -0.021 |
| pcnx2 | 0.196 | hmgb1a | -0.021 |
| uraha | 0.191 | uqcrc1 | -0.021 |
| camta1 | 0.187 | insm1a | -0.021 |