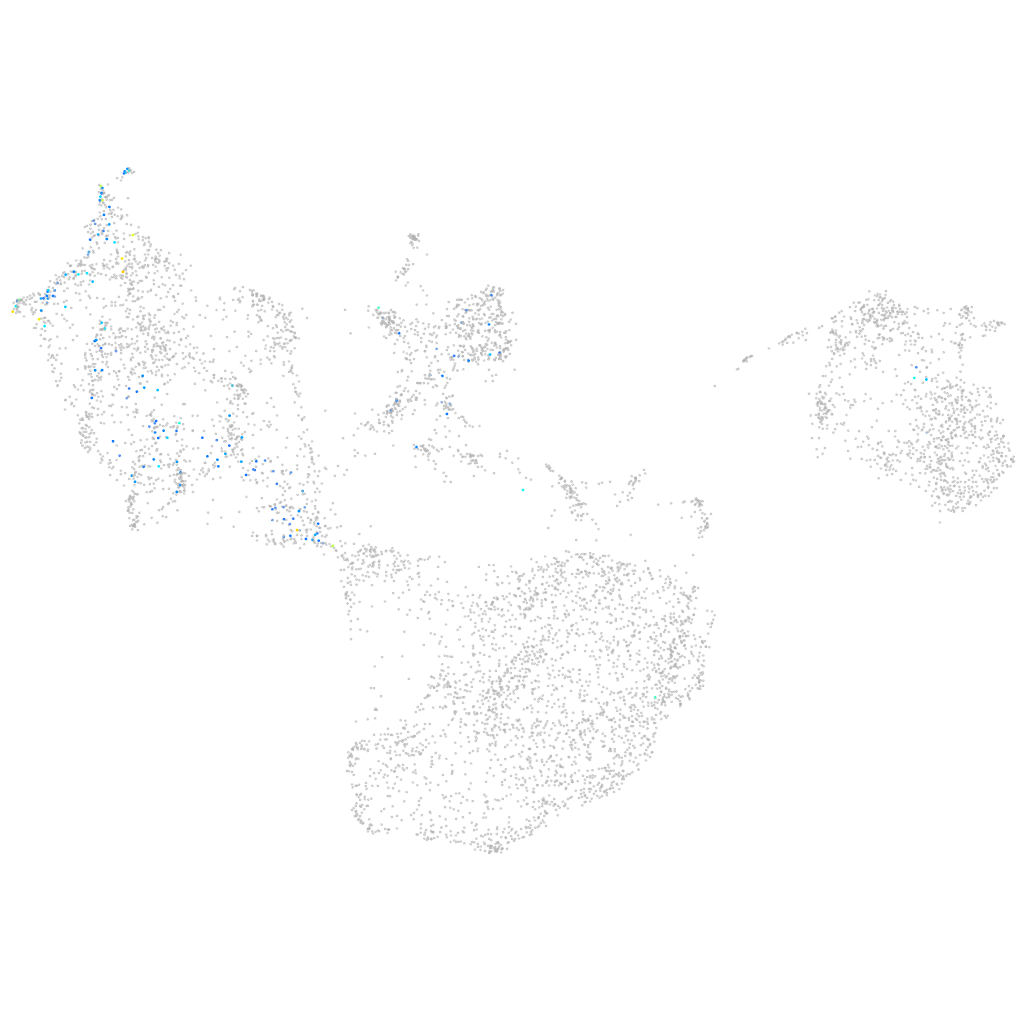
si:dkey-283j8.1
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle 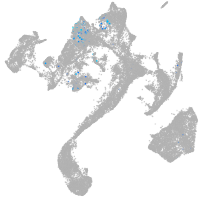 |
mural cells / non-skeletal muscle  |
mesenchyme 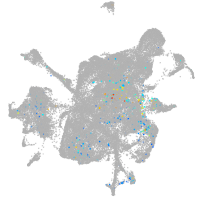 |
hematopoietic / vasculature 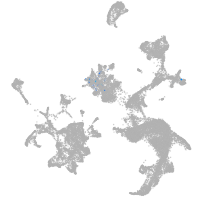 |
pronephros 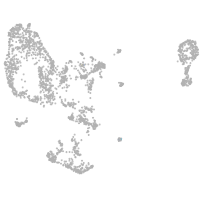 |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| vstm4a | 0.195 | cfl1l | -0.165 |
| tmem176 | 0.188 | cldni | -0.160 |
| cpn1 | 0.185 | tmsb1 | -0.158 |
| gpc1a | 0.185 | s100a10b | -0.157 |
| CU695073.1 | 0.183 | cldn1 | -0.155 |
| crabp2a | 0.181 | krt4 | -0.153 |
| tgfbi | 0.179 | pfn1 | -0.153 |
| cd82a | 0.178 | epcam | -0.152 |
| FP015860.1 | 0.176 | cotl1 | -0.151 |
| fah | 0.175 | cyt1 | -0.147 |
| si:ch211-286o17.1 | 0.171 | anxa1a | -0.145 |
| ptx3a | 0.171 | myh9a | -0.142 |
| sfrp1a | 0.169 | apoeb | -0.140 |
| lamc3 | 0.169 | spaca4l | -0.138 |
| fbln1 | 0.169 | bcam | -0.136 |
| msna | 0.165 | thy1 | -0.134 |
| agtr2 | 0.160 | arhgdig | -0.133 |
| pdgfra | 0.159 | tmsb4x | -0.133 |
| col11a1b | 0.158 | ecrg4b | -0.132 |
| mfap2 | 0.157 | rac2 | -0.132 |
| slc16a10 | 0.157 | si:dkey-248g15.3 | -0.132 |
| ankef1a | 0.157 | si:dkey-102c8.3 | -0.131 |
| fmoda | 0.157 | tagln2 | -0.130 |
| twist1b | 0.156 | col18a1a | -0.128 |
| ttc36 | 0.156 | egfl6 | -0.127 |
| inka1a | 0.155 | zgc:92380 | -0.127 |
| bhmt | 0.154 | rbp4 | -0.126 |
| tmem119b | 0.154 | hbegfa | -0.125 |
| cd81a | 0.152 | pycard | -0.124 |
| si:ch211-106j24.1 | 0.152 | arpc5b | -0.123 |
| marcksl1a | 0.151 | icn | -0.121 |
| dcn | 0.151 | tp63 | -0.121 |
| pmp22a | 0.151 | zgc:86896 | -0.120 |
| prrx1b | 0.151 | krt5 | -0.119 |
| pag1 | 0.150 | arhgef5 | -0.119 |