si:dkey-27h10.2
ZFIN 
Other cell groups


all cells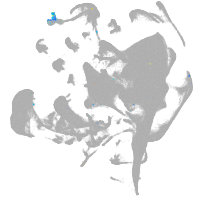 |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
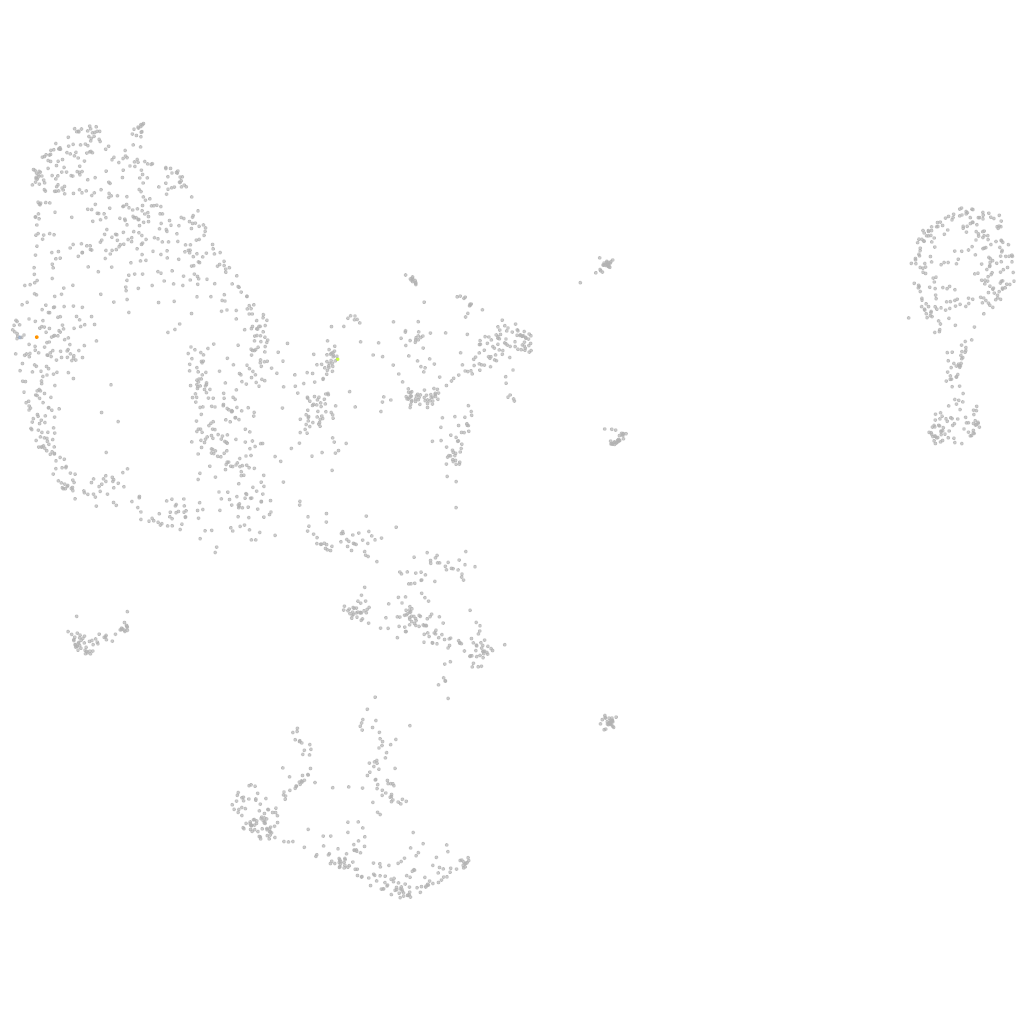
hematopoietic / vasculature 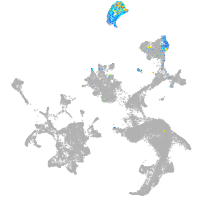 |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| sc:d156 | 0.570 | fkbp1aa | -0.077 |
| CU499336.2 | 0.563 | fthl27 | -0.072 |
| CABZ01001434.1 | 0.561 | ptmab | -0.062 |
| dok2 | 0.553 | atp5if1a | -0.058 |
| itln1 | 0.553 | CR383676.1 | -0.055 |
| FERMT3 (1 of many) | 0.548 | atp5mc3a | -0.055 |
| CABZ01050944.1 | 0.542 | mt-nd2 | -0.053 |
| LOC101884267 | 0.527 | ndufa6 | -0.053 |
| vaspa | 0.502 | tma7 | -0.053 |
| tnfaip8l2a | 0.470 | pnrc2 | -0.053 |
| npsn | 0.431 | si:ch73-1a9.3 | -0.053 |
| si:dkey-102g19.3 | 0.430 | cirbpa | -0.052 |
| serpina1l | 0.427 | atp5po | -0.052 |
| glipr1a | 0.410 | cirbpb | -0.051 |
| cpne4a | 0.408 | NC-002333.17 | -0.051 |
| pdcb | 0.407 | hnrnpaba | -0.050 |
| zdhhc3a | 0.395 | cox7c | -0.049 |
| spi1b | 0.395 | edf1 | -0.049 |
| rasal3 | 0.378 | atp5l | -0.048 |
| wasb | 0.376 | rbm8a | -0.048 |
| pkmb | 0.356 | calm2b | -0.048 |
| CU571069.1 | 0.354 | sem1 | -0.048 |
| ponzr6 | 0.338 | hmgn2 | -0.048 |
| grap2b | 0.335 | h3f3d | -0.048 |
| ela2 | 0.325 | ndufab1b | -0.048 |
| spice1 | 0.323 | hnrnpa0b | -0.048 |
| epd | 0.317 | ndufb10 | -0.048 |
| fcer1gl | 0.291 | ddx5 | -0.047 |
| zgc:111983 | 0.291 | cox6c | -0.047 |
| coro1a | 0.286 | h2afvb | -0.047 |
| CR376740.2 | 0.285 | cox5aa | -0.046 |
| lyz | 0.276 | ndufa8 | -0.046 |
| sult1st6 | 0.258 | ndufb7 | -0.046 |
| wipf1b | 0.246 | ndufb2 | -0.045 |
| BX088688.1 | 0.244 | eif2s3 | -0.044 |