si:dkey-27c15.3
ZFIN 
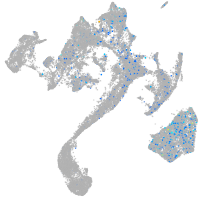
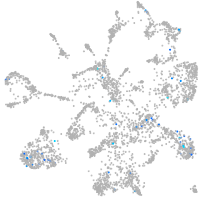
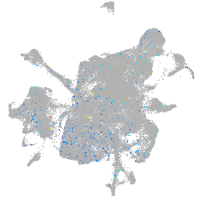
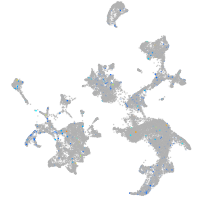
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| FP085399.4 | 0.076 | slc1a2b | -0.038 |
| BX324006.3 | 0.067 | ckbb | -0.036 |
| rrm1 | 0.065 | glula | -0.036 |
| LOC110437791 | 0.063 | cx43 | -0.035 |
| fbxo5 | 0.062 | mdkb | -0.034 |
| XLOC-021212 | 0.062 | atp1a1b | -0.034 |
| lbr | 0.060 | slc6a1b | -0.033 |
| ssuh2.3 | 0.060 | efhd1 | -0.033 |
| dut | 0.058 | acbd7 | -0.033 |
| mki67 | 0.058 | pvalb2 | -0.032 |
| stmn1a | 0.057 | slc3a2a | -0.032 |
| anp32b | 0.057 | pvalb1 | -0.032 |
| rrm2 | 0.056 | slc4a4a | -0.031 |
| zgc:165555.3 | 0.056 | mt2 | -0.030 |
| LOC103909971.1 | 0.055 | ppap2d | -0.030 |
| CR352218.1 | 0.055 | fabp7a | -0.030 |
| pcna | 0.055 | si:ch211-66e2.5 | -0.030 |
| si:dkeyp-114g9.1 | 0.054 | ptn | -0.030 |
| CABZ01005379.1 | 0.054 | hepacama | -0.029 |
| chaf1a | 0.054 | gapdhs | -0.028 |
| dek | 0.054 | mgll | -0.028 |
| lig1 | 0.053 | gpr37l1b | -0.027 |
| hmga1a | 0.053 | mfge8a | -0.027 |
| dhfr | 0.053 | ak1 | -0.026 |
| mcm7 | 0.053 | cdo1 | -0.026 |
| cks1b | 0.053 | sept8b | -0.026 |
| h3f3a | 0.053 | cspg5b | -0.026 |
| ccna2 | 0.053 | aldocb | -0.025 |
| seta | 0.053 | CU467822.1 | -0.025 |
| ranbp1 | 0.053 | sema6a | -0.025 |
| C1orf232 | 0.053 | FO704813.1 | -0.025 |
| aurkb | 0.053 | si:ch1073-303k11.2 | -0.025 |
| zgc:110540 | 0.052 | id2a | -0.025 |
| rpa2 | 0.052 | proza | -0.024 |
| chek1 | 0.051 | cox4i2 | -0.024 |