si:dkey-271j15.3
ZFIN 
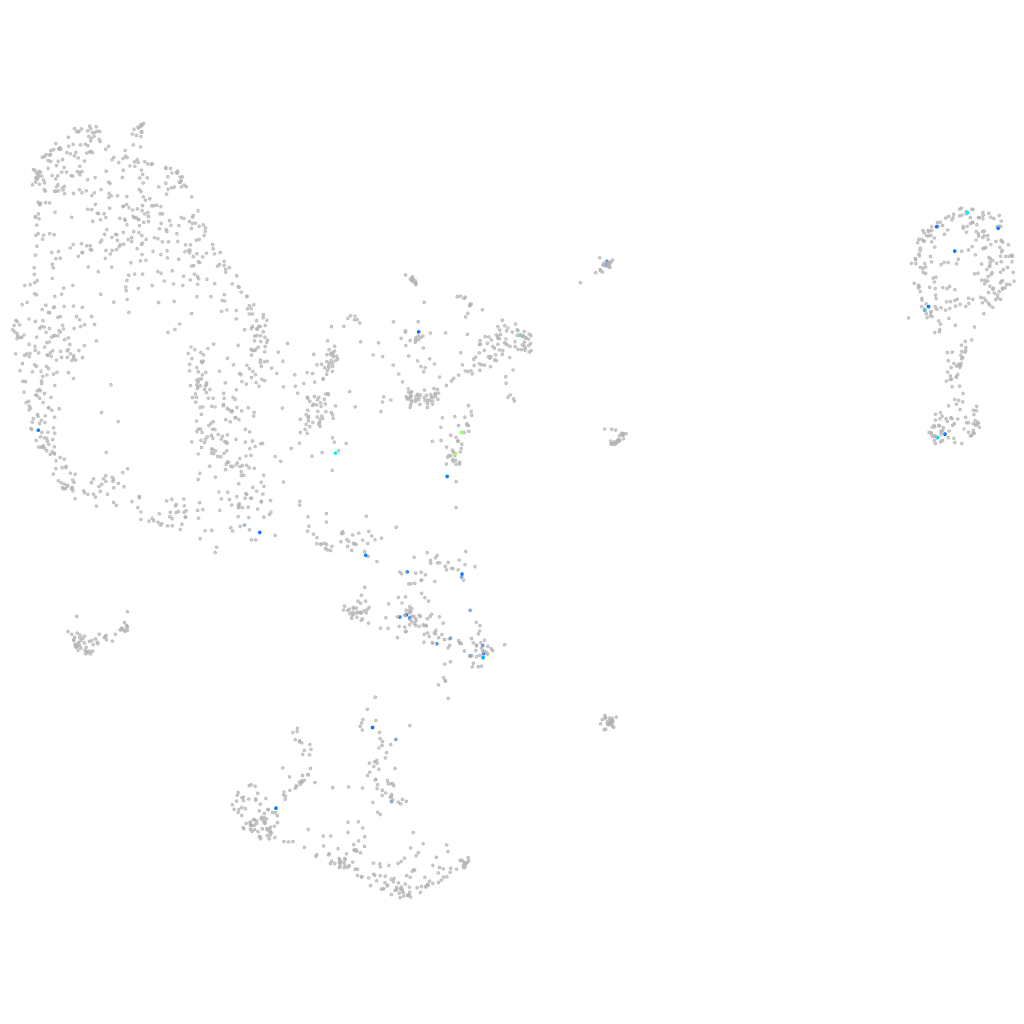
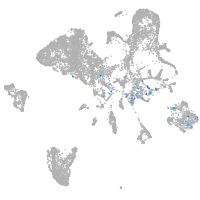
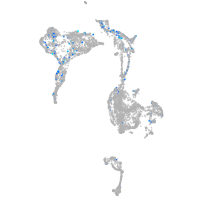
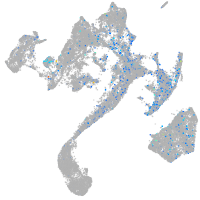
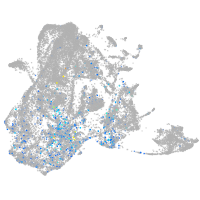
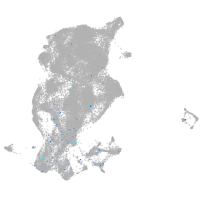
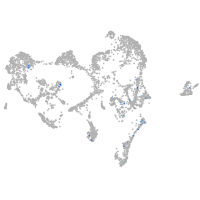
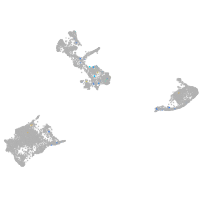
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CU467822.2 | 0.333 | cdaa | -0.096 |
| marco | 0.333 | gstr | -0.096 |
| zgc:92162 | 0.333 | si:ch211-139a5.9 | -0.095 |
| lrrc3ca | 0.327 | rpl34 | -0.095 |
| mb21d2 | 0.303 | COX3 | -0.095 |
| malrd1 | 0.303 | aldh6a1 | -0.094 |
| si:ch211-215p11.1 | 0.297 | vdac3 | -0.093 |
| dip2bb | 0.293 | suclg1 | -0.093 |
| pcdh1g22 | 0.286 | elovl1b | -0.093 |
| dicp1.19 | 0.283 | cdh17 | -0.091 |
| XLOC-042136 | 0.283 | cox5ab | -0.091 |
| XLOC-044118 | 0.283 | aldh7a1 | -0.090 |
| LOC101883659 | 0.280 | si:dkey-16p21.8 | -0.090 |
| irx5a | 0.277 | mt-atp6 | -0.089 |
| mpp4a | 0.276 | mt-co2 | -0.089 |
| CR318601.1 | 0.276 | atp5l | -0.088 |
| cmklr1 | 0.266 | ahcyl1 | -0.088 |
| CABZ01016118.1 | 0.262 | atp1a1a.4 | -0.088 |
| SAMD5 | 0.260 | mt-co1 | -0.088 |
| cdnf | 0.257 | gpd1b | -0.087 |
| fsip1.1 | 0.251 | mt-nd4 | -0.087 |
| XLOC-011956 | 0.251 | nit2 | -0.087 |
| LOC100330501 | 0.250 | sgk1 | -0.087 |
| LO018196.1 | 0.248 | ndufa1 | -0.086 |
| prelid3a | 0.245 | dera | -0.086 |
| mfng | 0.244 | gstp1 | -0.085 |
| crfb16 | 0.242 | gcshb | -0.085 |
| LOC101886621 | 0.238 | ahcy | -0.084 |
| dicp3.1 | 0.238 | agxta | -0.084 |
| FP085421.1 | 0.237 | itm2bb | -0.083 |
| kctd20 | 0.234 | bsg | -0.083 |
| ptprna | 0.230 | suclg2 | -0.083 |
| CABZ01080074.1 | 0.227 | atp5mc3b | -0.083 |
| cyp7b1 | 0.226 | mdh1aa | -0.082 |
| cth1 | 0.224 | pnp6 | -0.082 |